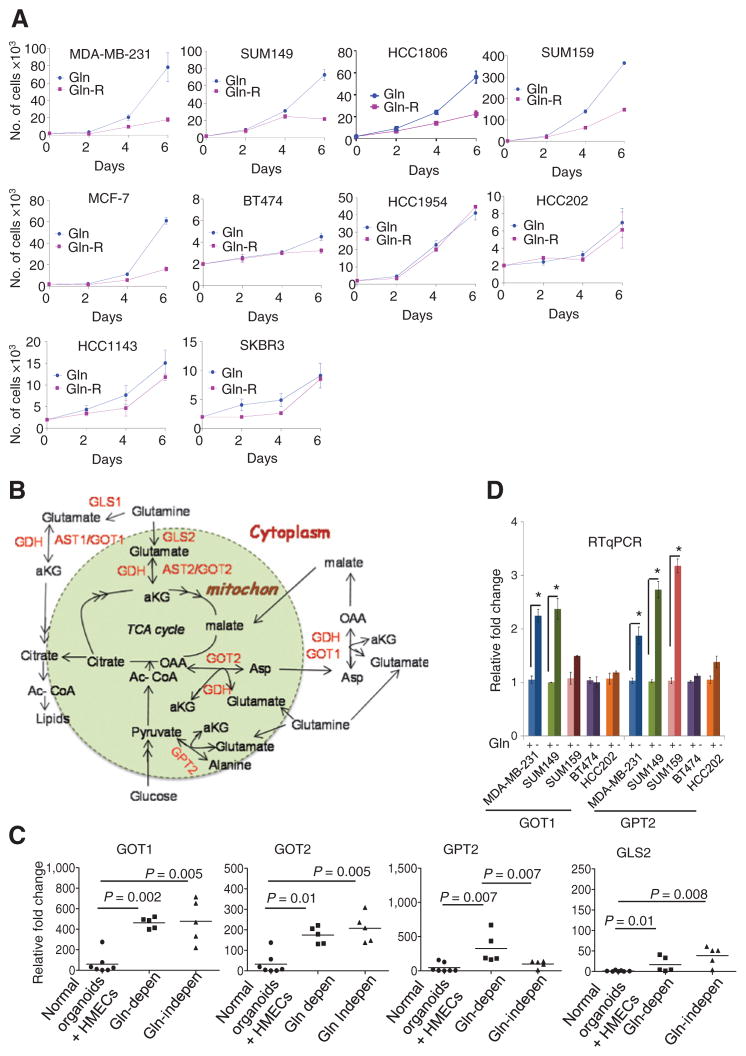
Figure 1.
Glutamine dependency of breast cancer cell lines. A, breast cell lines were grown in media with complete or reduced glutamine (Gln and Gln-R). The average number of live cells per well (3-wells/time point; two replicates), on indicated days is shown. B, schematic representation of transaminases and other genes involved in glutaminolysis. C, differential mRNA expression of GOT1, GOT2, GLS2, and GPT2 in breast cancer cell lines compared with normal mammary organoids and HMECs (duplicates in each assay; three replicates). The Mann–Whitney test, *, P < 0.05. D, RT-qPCR of genes in the glutaminolytic pathway following growth in medium with complete (Gln+) or reduced glutamine (Gln−) for 24 hours (duplicates in each assay; three replicates); the Mann–Whitney test; *, P < 0.05.