Fig. 1.
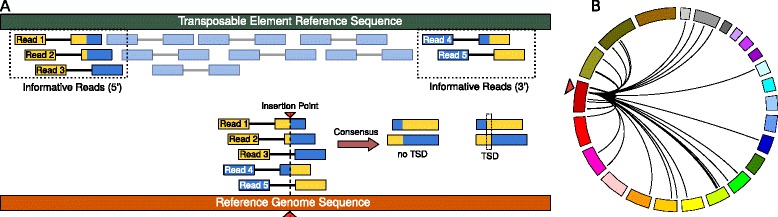
Read mapping patterns typically associated with insertion detection. Panel a shows the read mapping patterns versus a reference TE sequence (grey rectangle, top) and the mapping of the same reads to a reference genome sequence (orange rectangle, bottom). Reads are represented as typical paired-end reads where the ends of each amplicon are represented as rectangles and the un-sequenced portion of the amplicons are represented as bars connecting the rectangles. Reads informative for identifying TE insertion locations are indicated by dashed boxes, other read mappings to the TE reference are shown in light blue boxes. Within the informative reads, reads or portions of reads mapping to the TE reference are coloured blue, and mappings to the reference genome sequence are coloured yellow. The exact location of this example insertion is indicated by the red triangle and the dashed line. Assembly of the reads supporting the two junction sequences is indicated to the right of the ‘consensus’ arrow, one example with a TSD and one without. If a TSD is present, the insertion breakends relative to the reference genome are staggered, and the overlap of reference-aligned sequence corresponds to the TSD. If a TSD is not present (and no bases are deleted upon insertion), the junctions obtained from the 5' end and the 3' end of the TE reference will match exactly. Panel b shows a typical pattern of discordant read mappings across a genome - the colored segments in circle represent chromosomes, each black link indicates a discordant read mapping supporting an insertion at the position indicated by the red triangle. The endpoints not corresponding to the insertion site map to TE elements at various locations in the reference genome
