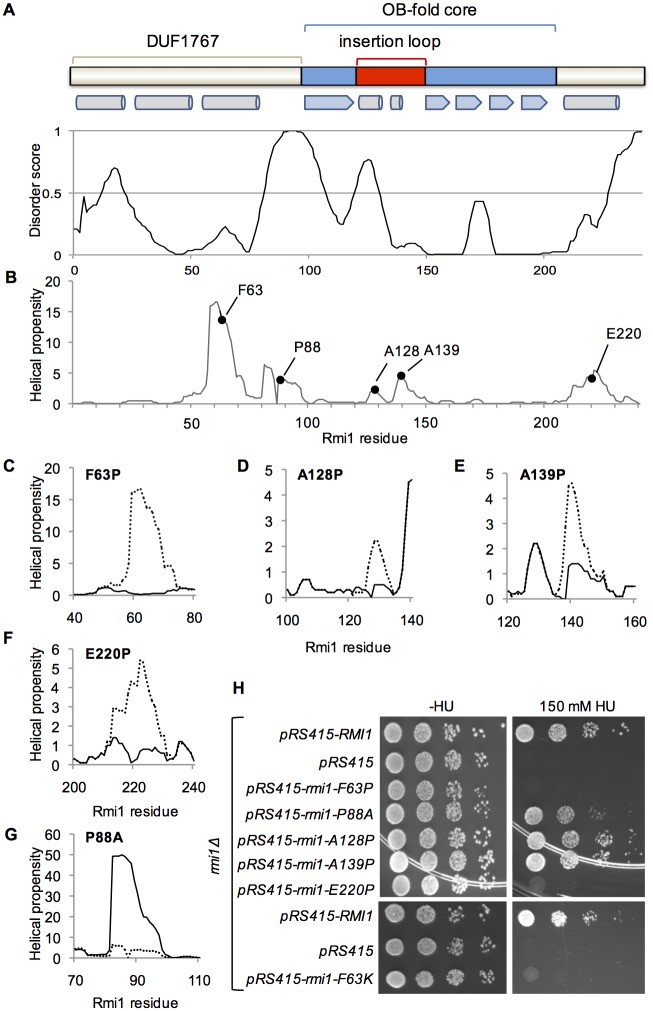
Fig 1. Structure-prediction-guided mutagenesis of S.c. Rmi1.
A, prediction of order/disorder in Rmi1 by the VLXT algorithm [26,27]. A score of 1 denotes an ideal prediction of disorder and a score of 0 an ideal prediction of order with the order/disorder threshold at a score of 0.5. A domain of unknown function (DUF1767), and an OB-fold with an insertion loop are conserved in all Rmi1 species [39]. The position of DUF1767, the OB-fold, the insertion loop and a predicted flexible linker between DUF1767 and the OB-fold shown above the disorder plot are based on the VLXT order/disorder prediction. α-helices (cylinders) and β-strands (arrows) in this region of human Rmi1 are indicated below the domain map. B, prediction of four regions of increased helical in Rmi1 with residues F63, A128,A139, E220 having some of the highest helical propensity in the DUF1767 domain, the insertion loop, and the C-terminus, respectively. We predict that P88 is responsible for the sudden loss in helical propensity in the linker that connects N-terminal domain and the OB-fold. C–G, substitution of F63, A128, A139 and E220 with proline, which has the lowest helical propensity of all amino acids, is predicted to disrupt the increased helical propensity in these regions, whereas substitution of P88 with alanine, which has excellent helical propensity, is predicted to lead to a strong increase in continuous helical propensity of the linker. H, plasmid pRS415 expressing RMI1 and rmi1 mutants under control of the endogenous RMI1 promoter were transformed into Δrmi1 mutant KHSY4695 and tested for the ability to suppress the hypersensitivity of the rmi1Δ strain to hydroxyurea.