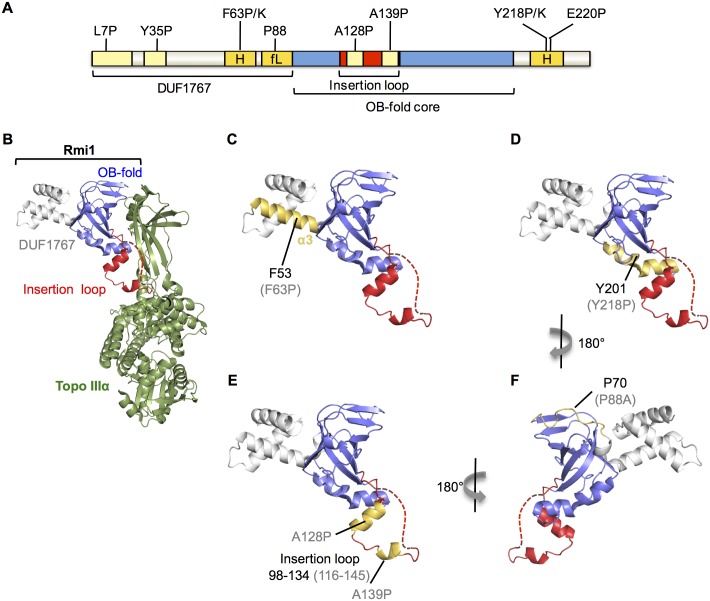
Fig 6. Conserved domains, and putative differences between yeast Rmi1 and the N-terminus of human Rmi1.
A, functional importance of seven structural motifs predicted in yeast Rmi1 was tested by analyzing point mutations in vivo. Mutation of putative motifs highlighted in light yellow did not impair Rmi1 function in vivo, including L7P and Y35P mutations in the putative DUF1767 domain, and A128P and A139P in the topoisomerase-binding loop. Mutations in motifs highlighted in dark yellow impaired Rmi1 function in vivo; F63P and F63K mutations in an N-terminal α-helix (H), Y218P, Y218K, and E220P mutations in a C-terminal α-helix (H) caused null phenotypes, and the P88A mutation in the flexible linker (fL) between the DUF1767 domain and the OB-fold caused intermediate functional impairment. B, he crystal structure of the N-terminus of human Rmi1 bound to Topo IIIα (4CGY [19]) was rendered in PyMol. The DUF1767 domain, the OB-fold core and the insertion loop extending between strands β1 and β2 are shown in grey, blue, and red, respectively. A part of the disordered insertion loop missing from the crystal structure is indicated by a red dashed line. Topo IIIα is shown in green. C, the domain structure of Rmi1 is shown as in (B) with α3, corresponding to the functionally important, putative α-helix in yeast Rmi1, highlighted in yellow. Residue F53 of human Rmi1 and the corresponding F63P null mutation in yeast Rmi1 (in brackets) are indicated. D, the C-terminal α-helix interacting with the bottom of the OB-fold core is shown in yellow. Y201 and the corresponding Y218P null mutation in yeast Rmi1 (in parentheses) are indicated. Disruption of this α-helix leads to lower Rmi1 expression levels and total loss of Rmi1 function (Fig 3E). E, human Rmi1 contains two helical segments in the insertion loop, shown in yellow. A128P and A139P mutations were designed to disrupt helical propensity in the corresponding insertion loop in yeast Rmi1, but did not interrupt Rmi1 function. F, a flexible linker connects the DUF1767 domain to the OB-fold in human Rmi1, indicated in yellow. A single proline at position 88 is predicted to disrupt the strong helical propensity of this linker in yeast Rmi1 whereas this linker is more proline-rich in human Rmi1. The location of P70 in human Rmi1, corresponding to P88 in yeast Rmi1, is indicated.