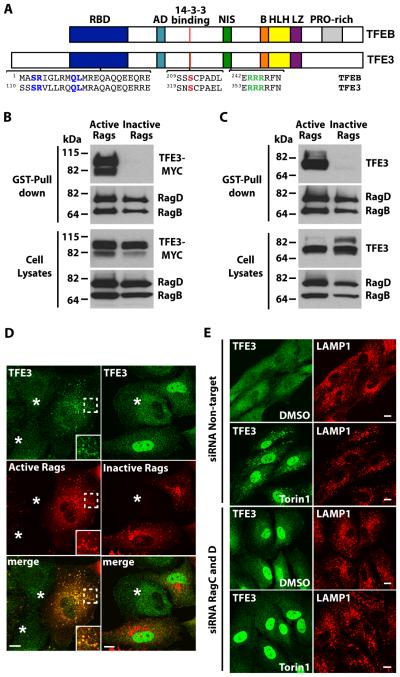
Fig. 2. TFE3 interacts with active Rag heterodimers.
(A) Schematic representation of comparison between TFEB and TFE3. RBD: Rag-binding domain, AD: Activation domain, NIS: Nuclear import signal, B: Basic residues, HLH: Helix-loop-helix, LZ: Leucine-zipper, PRO-rich: Proline rich. Residues proven to be essential in the RBD (S112R113), 14-3-3 binding site (S321), and NIS (R356R357R358) are colored. (B and C) Immunoblotting analysis of TFE3-MYC (B) or endogenous TFE3 (C) affinity purified with glutathione S-transferase (GST) fused to C-terminus of active Rag heterodimers (RagBGTP/RagDGDP) in ARPE-19 cells. The affinity-purified materials were probed with antibodies against GST, MYC, and TFE3 (used to detect Rag proteins, TFE3-MYC, and endogenous TFE3, respectively). Data are representative of 3 independent experiments. (D) Immunofluorescence confocal microscopy analysis of the subcellular distribution of endogenous TFE3 in ARPE-19 cells overexpressing either active or inactive RagB/D heterodimers. Cells were double stained with antibodies against TFE3 and GST (used to detect endogenous TFE3 or Rag proteins, respectively). Asterisks indicate distribution of endogenous TFE3 in non-transfected cells. Scale bars, 10 μm Data are representative of 3 independent experiments and over 90% of cells positive for the RagB/D heterodimers exhibited the phenotypes shown. (E) Immunofluorescence confocal microscopy analysis of the subcellular distribution of endogenous TFE3 in control or Rag-depleted ARPE-19 cells upon incubation with Torin-1 for 1 hour. Cells were double stained with antibodies against TFE3 and LAMP1. Scale bars, 10 μm. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown.