FIG. 2.
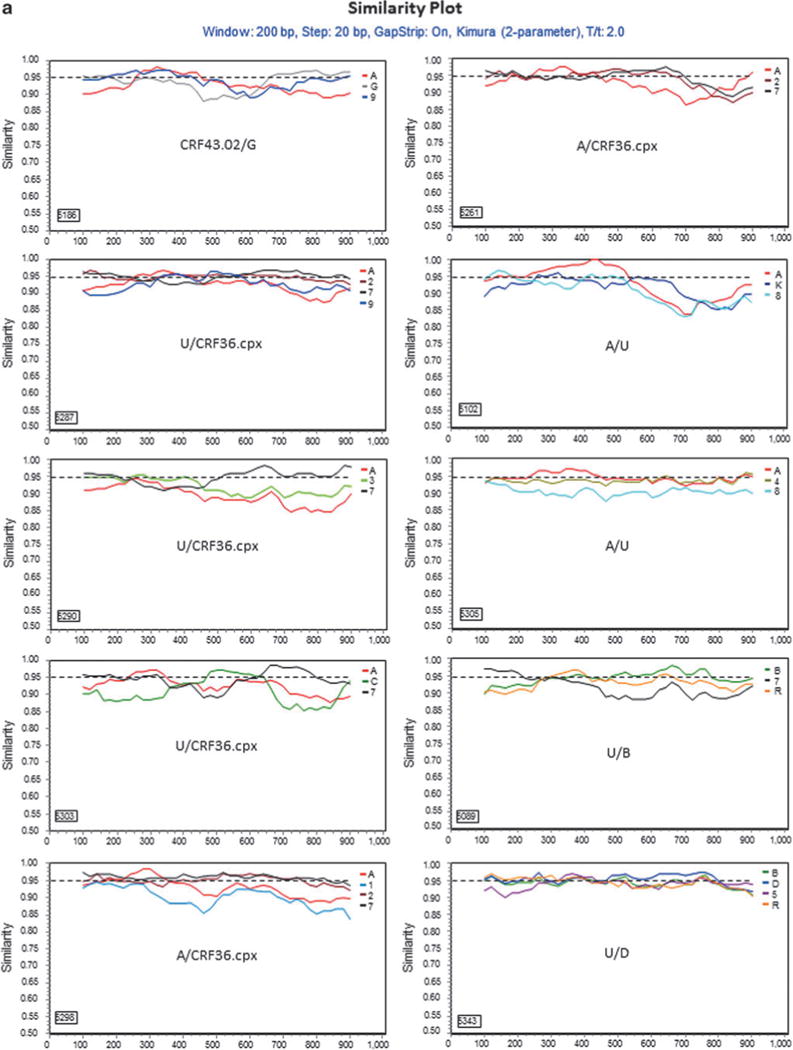
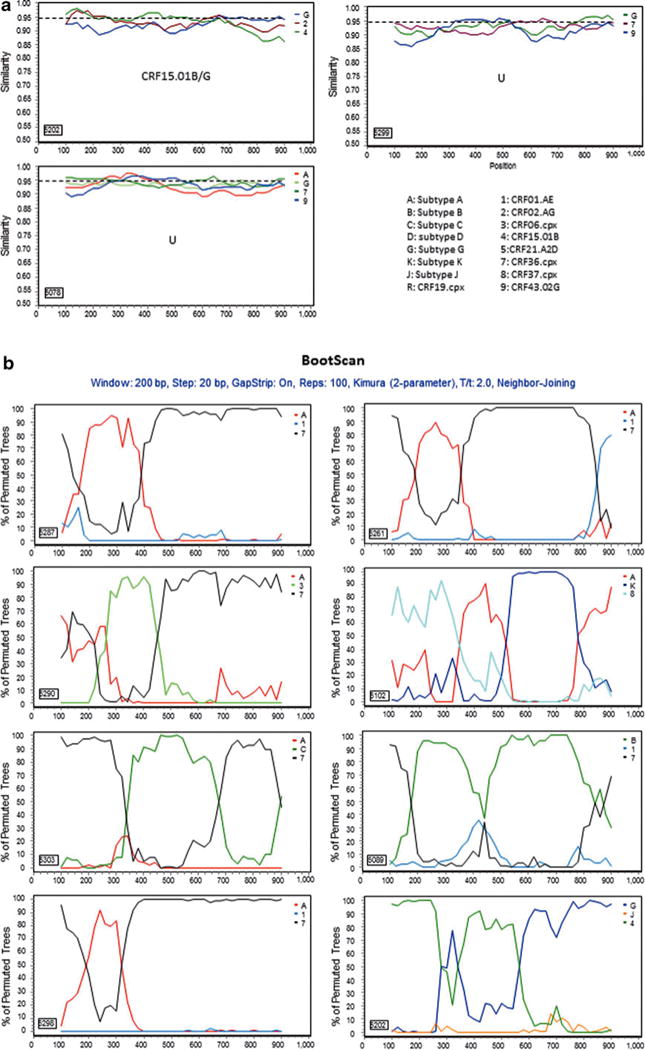
Further characterization of phylogenetically unresolved Nigerian sequences. The 13 representative Nigerian unclassifiable sequences (Table 1) were further analyzed individually using SimPlot59 for genetic similarity (200-bp window, 20-bp step, and two-parameter Kimura algorithm). The selection of HIV-1 reference subtypes for analysis was based on the genetic distance analysis (Table 2). Only three to four relevant reference subtypes were chosen to generate the graph to show the gene structure of query sequences (a). The dashed line was used to determine the relatedness of the query sequence to the selected reference subtypes at the 95% similarity threshold across the gene. The segment of the query sequence with a length of less than 200 nucleotides and less than 95% similarity to the references was assigned a “U” indicating an unresolved subtyping for the sequence portion. The chimeric subtyping of the query sequence was given in each of the SimPlot diagrams (a) and populated in Table 2. To explore the recombinant events, 8 among the 13 unclassifiable (U) sequences were again analyzed using the BootScan59 method, demonstrating a clear recombination event (b).
