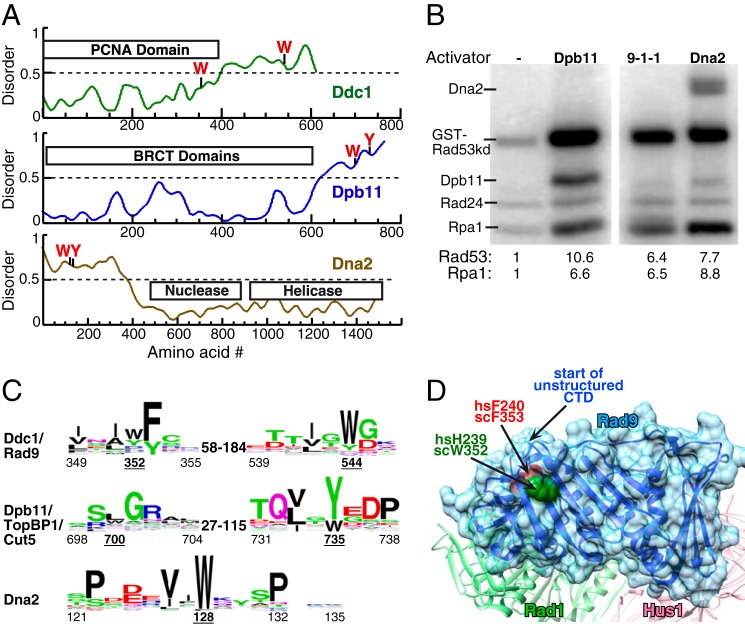
FIGURE 1.
Three activators of S. cerevisiae Mec1. A, schematic presentation of the domain structure of Ddc1, Dpb11, and Dna2 of S. cerevisiae. The aromatic residues required for Mec1 activation are indicated. Structural disorder was calculated using IUPRED (40). Regions with disorder parameter >0.5 are considered to be intrinsically disordered. B, kinase assay comparing the ability of 20 nm full-length Dpb11, 9-1-1, or Dna2 to activate Mec1. Mec1 stimulation over basal activity is indicated as fold-activation for either GST-Rad53-kd or Rpa1; see “Experimental Procedures” for details. C, WEB logos derived from sequence comparisons as described in “Experimental Procedures.” The range of distances between conserved motifs in different organisms is shown. Numbering below motifs is for the S. cerevisiae proteins with the underlined numbers indicating the critical aromatics. D, structure of the Rad9 subunit in human 9-1-1 from (27), with comparable human and S. cerevisiae residues indicated. The start of the unstructured C-terminal tail is also indicated.