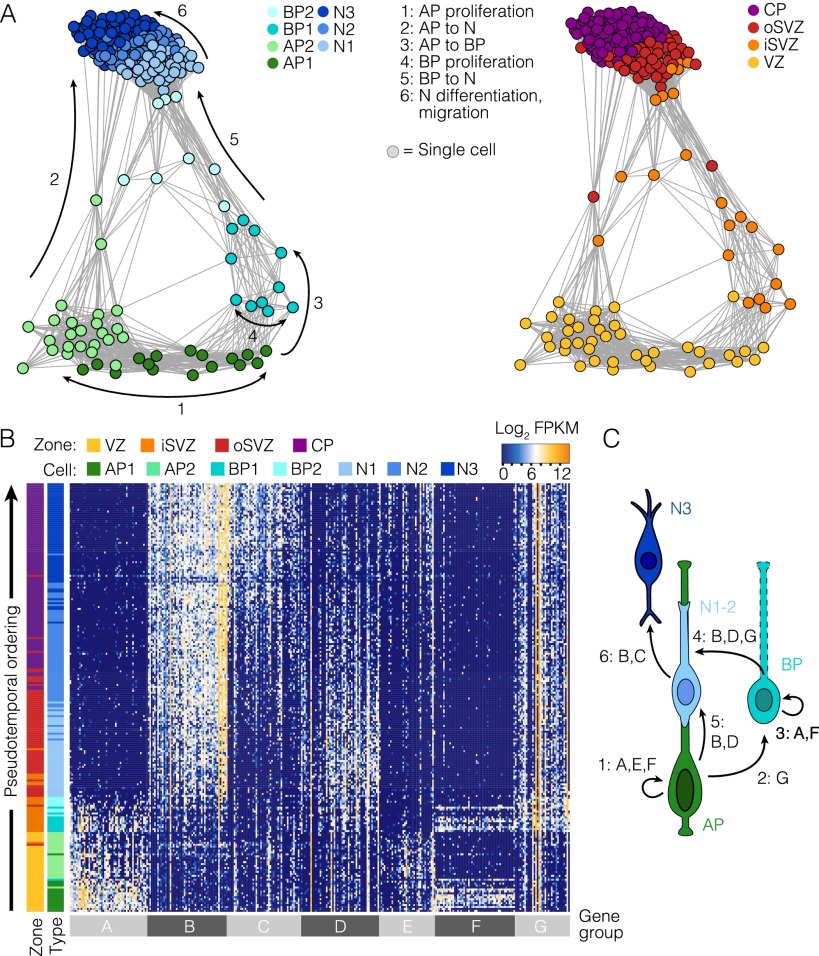
Fig. S2.
Gene expression signatures controlling fetal corticogenesis. (A) Cell lineage network based on pairwise correlations between cells and using the same gene set used in the Monocle analysis (Fig. 2 A and B). Cells are color-coded based on cell-type classification (Left) or maximum correlation to cortical zones (Right). Arrows and numbers mark topological features of the network that reflect cell proliferation and differentiation events inferred from gene expression signatures. (B) Ordering of scRNA-seq expression data according to the pseudotemporal position along the lineage revealed a continuum of gene expression changes from NPCs to neurons. Genes from the heat map depicted in Fig. 1C are shown. Each row represents a single cell and each column a gene. The cell-type assignment and maximum correlation to cortical zone are shown as sidebars. (C) Schematic illustrating lineage decisions in the fetal cortex, labeled with groups of genes characterizing lineage progression. For example, APs are defined by the expression of group A genes and self-renew through the expression of cell-cycle regulators shown in gene groups E and F.