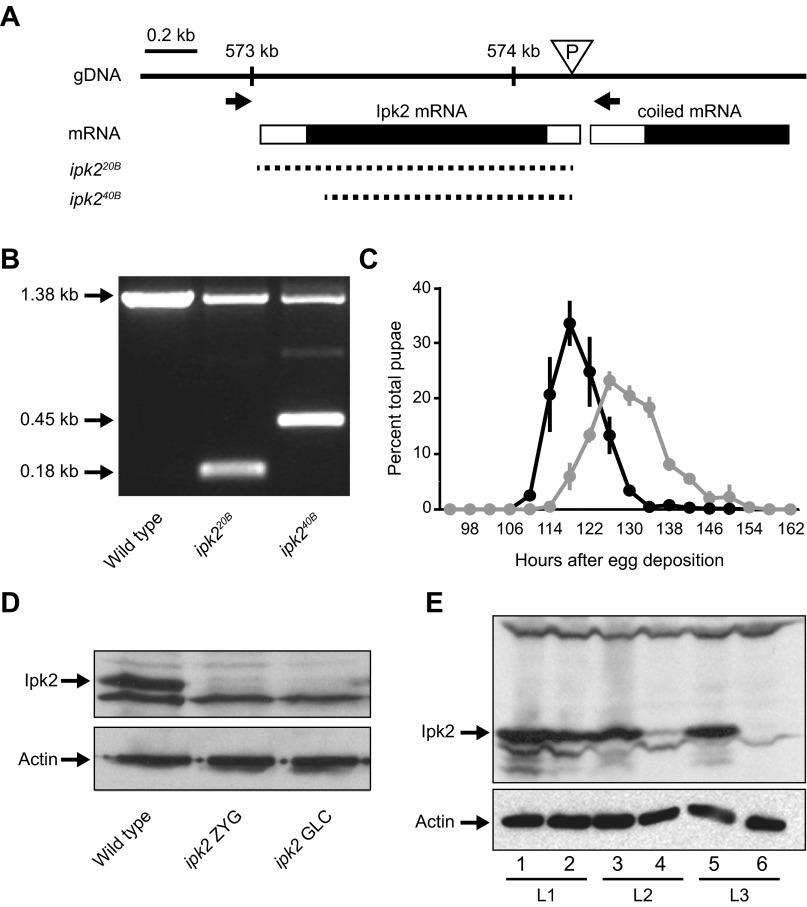
Fig. S1.
ipk2 excision mutants. (A) Schematic representation of the Ipk2 locus on the second chromosome (2L). The triangle represents the location of the ipk2G3545 P-element in the 5′ untranslated region that was used to generate the excision lines. Arrows show the locations of primers used to screen for unidirectional excisions. Dashed lines represent the missing genomic DNA in the excision or deficiency lines. ipk220B and ipk240B are the alleles generated in this study. (B) Single fly PCR of wild-type, ipk220B, and ipk240B heterozygous alleles using the primers indicated in A. The excision boundaries (shown in A) were determined based on sequence analysis of the PCR products. The 1.38-kb band indicates a wild-type ORF, whereas the 0.45- and 0.18-kb bands represent the shortened ORFs of the mutants. (C) Delayed pupation of the homozygous ipk220B mutant. The percent of total animals that pupated at each time point is plotted (mean ± SEM). (D) Western blot of ipk2 zygotic (ipk2 ZYG) and germ-line clone (ipk2 GLC) wandering third-instar larval mutants. (E) Perdurance of maternal Ipk2 protein during larval development in ipk2 zygotic mutants. Protein lysates from stage L1 (lanes 1 and 2), L2 (lanes 3 and 4), and L3 (lanes 5 and 6) of wild type (lanes 1, 3, and 5) and ipk2 (lanes 2, 4, and 6) were subject to SDS/PAGE and immunoblotted for Ipk2p (Upper). Compared with wild-type L1 larvae, L1 ipk2 null larvae have comparable levels of Ipk2 protein, whereas it drops to a lower level in L2 and is undetectable in L3.