Abstract
Epigenetic modifiers have fundamental roles in defining unique cellular identity through the establishment and maintenance of lineage-specific chromatin and methylation status1. Several DNA modifications such as 5-hydroxymethylcytosine (5hmC) are catalysed by the ten eleven translocation (Tet) methylcytosine dioxygenase family members2, and the roles of Tet proteins in regulating chromatin architecture and gene transcription independently of DNA methylation have been gradually uncovered3. However, the regulation of immunity and inflammation by Tet proteins independent of their role in modulating DNA methylation remains largely unknown. Here we show that Tet2 selectively mediates active repression of interleukin-6 (IL-6) transcription during inflammation resolution in innate myeloid cells, including dendritic cells and macrophages. Loss of Tet2 resulted in the upregulation of several inflammatory mediators, including IL-6, at late phase during the response to lipopolysaccharide challenge. Tet2-deficient mice were more susceptible to endotoxin shock and dextran-sulfate-sodium-induced colitis, displaying a more severe inflammatory phenotype and increased IL-6 production compared to wild-type mice. IκBζ, an IL-6-specific transcription factor, mediated specific targeting of Tet2 to the Il6 promoter, further indicating opposite regulatory roles of IκBζ at initial and resolution phases of inflammation. For the repression mechanism, independent of DNA methylation and hydroxymethylation, Tet2 recruited Hdac2 and repressed transcription of Il6 via histone deacetylation. We provide mechanistic evidence for the gene-specific transcription repression activity of Tet2 via histone deacetylation and for the prevention of constant transcription activation at the chromatin level for resolving inflammation.
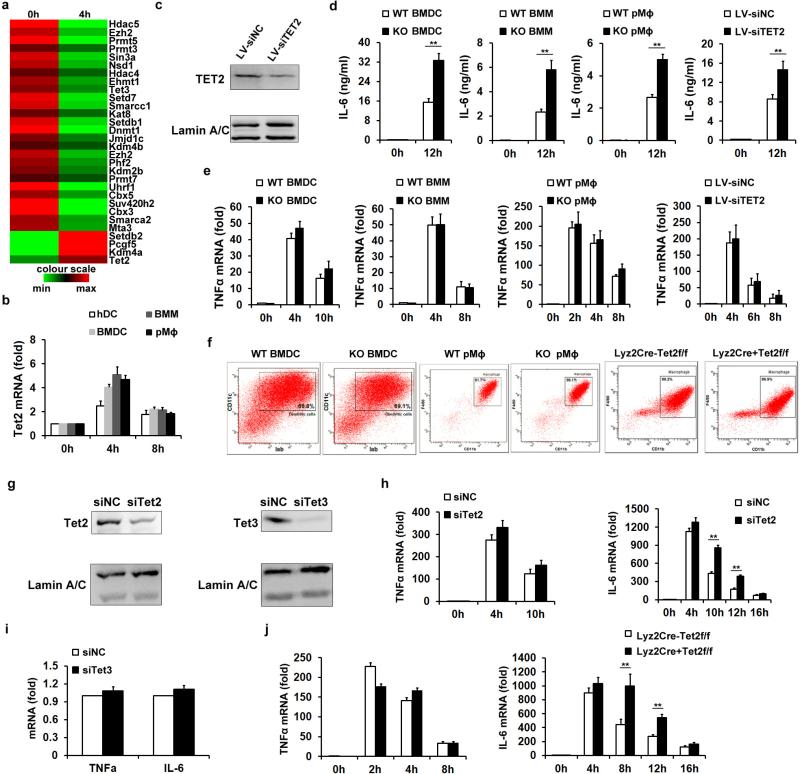
Tet proteins mediate de novo 5-hmC in mammals and play important roles in transcription regulation2,4. Dynamic transcriptomes of murine bone marrow-derived dendritic cells (BMDC) showed upregulation of Tet2 and downregulation of Tet3 among chromatin modifiers during lipopolysaccharide (LPS) response (Extended Data Fig. 1a and Supplementary Table 1). Transcription of Tet2 also increased in both human and murine innate myeloid cells after LPS stimulation (Extended Data Fig. 1b). Although Tet2 is an intrinsic repressor of myeloid leukaemia5, the roles of Tet2 in innate immunity and inflammation are unknown.
Using Tet2-deficient BMDC and macrophages and Tet2-silenced human dendritic cells, we found deficiency or knockdown of Tet2 did not significantly affect the marked increase of IL-6 messenger RNA expression at early phase of LPS stimulation (before 4 h). However, deficiency or knockdown of Tet2 led to Il6 mRNA expression to be maintained at higher levels at late phase (after 8 h) when mRNA levels of Il6 decreased significantly in control cells (Fig. 1a, b and Extended Data Fig. 1c). Accordingly, IL-6 protein levels also increased in these tested cells (Extended Data Fig. 1d). In contrast, Tnfa mRNA levels were similar at early and late phase of LPS stimulation in the analysed samples (Extended Data Fig. 1e). Developmental defects of dendritic cells and macrophages were not observed in Tet2-deficient mice (Extended Data Fig. 1f). We further confirmed the regulatory role of Tet2 in transcription of Il6 but not Tnf in Tet2-silenced macrophages (Extended Data Fig. 1g, h). Silencing of Tet3 barely altered Il6 transcription (Extended Data Fig. 1g, i), indicating Tet3 might not be involved in this process.
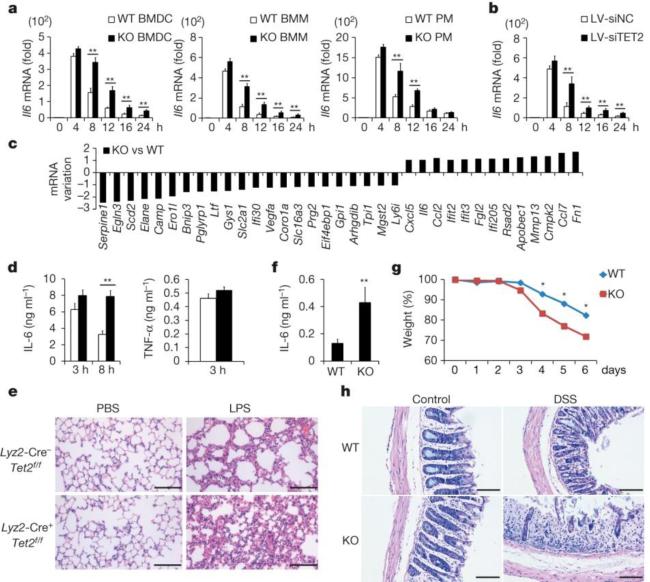
Figure 1. Loss of Tet2 maintains higher expression of IL-6.
a, b, Il6 mRNA in BMDC, bone-marrow-derived macrophages (BMM) and peritoneal macrophages (PM) (a) from wild-type (WT) and Tet2-knockout (KO) mice, and TET2-silenced human dendritic cells (b) during LPS response. c, log2 ratio of mRNA variations in Tet2-deficient BMDC 8 h after LPS stimulation. d, e, ELISA of sera cytokines (d) and histopathology of lungs (e) from conditional Tet2-deficient and control mice (n = 5) after intra-peritoneal injection of LPS (10 mg per kg body weight). Scale bars, 50 μm. f, g, h, ELISA of sera IL-6 (f), changes of body weights (g) and histopathology of colonic sections (h) of Tet2-deficient and control mice (n = 5) on day 6 after treatment with 3% DSS. Scale bars, 100 μm. Error bars represent s.d. of triplicate technical replicates (b, d, f) or s.e.m. of triplicate biological replicates (a). Data are representative of 3 independent experiments. Unpaired Student's t-test, *P < 0.05, **P < 0.01.
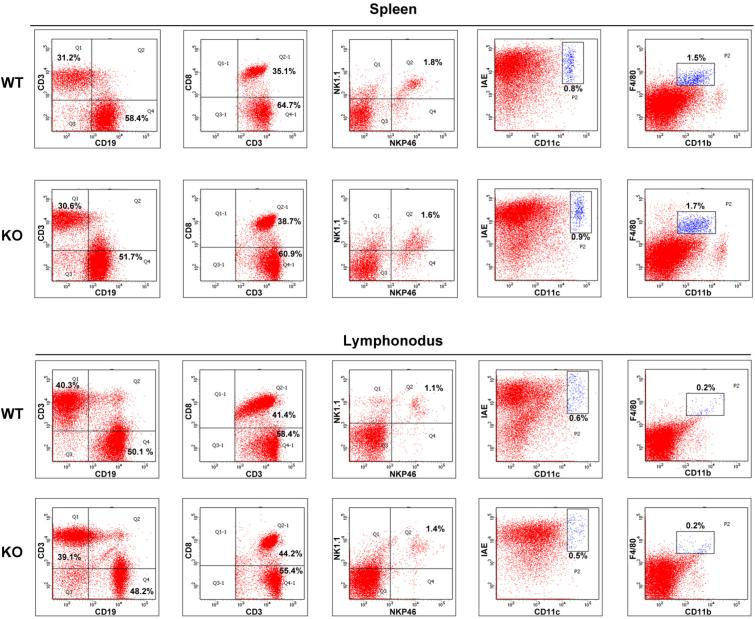
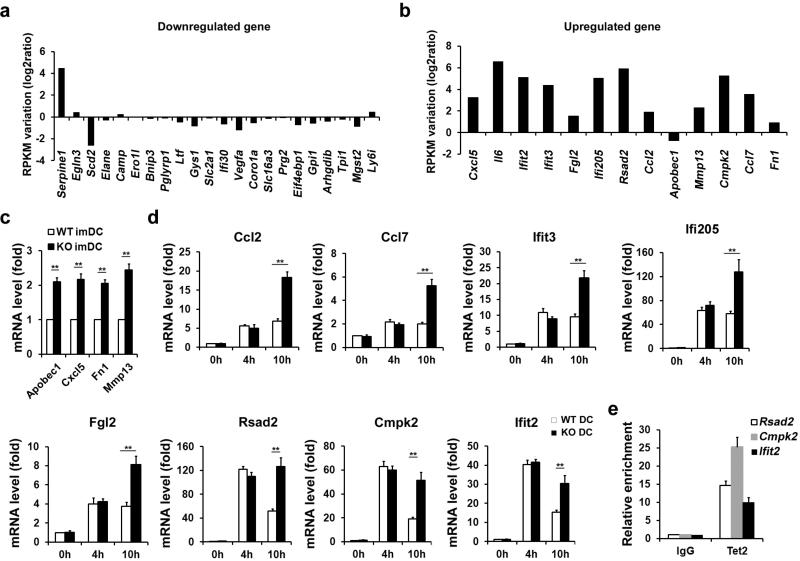
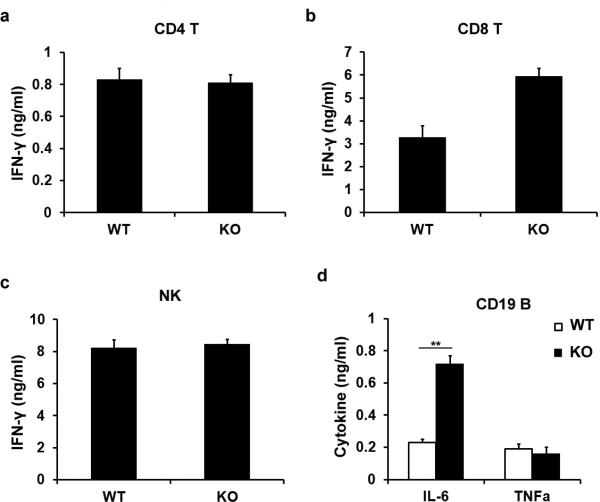
To broaden the role of Tet2 in repressing transcription of other proinflammatory genes like IL-6 during immune activation, we screened cytokine expression in both innate and adaptive immune response. Developmental defects of innate and adaptive immune cells were not observed in Tet2-deficient mice (Extended Data Fig. 2). We identified 22 downregulated genes and 13 upregulated genes in Tet2-deficient BMDC (Fig. 1c). We further analysed their dynamic expression patterns in RNA sequencing (RNA-seq) data of LPS-stimulated BMDC (Supplementary Table 1). For the genes downregulated in Tet2-deficient BMDC, we found that expression of these genes were mostly repressed or not affected by LPS, suggesting a different regulatory role of Tet2 for Toll-like receptor (TLR) signal-repressed or -unaffected genes (Extended Data Fig. 3a). For the genes upregulated owing to loss of Tet2, we found that 12 genes were TLR signal-induced genes, like Il6 (Extended Data Fig. 3b). Among the upregulated genes, Apobec1, Cxcl5, Fn1 and Mmp13 were upregulated in Tet2-deficient immature dendritic cells, suggesting a TLR signal-independent mechanism for Tet2-mediated regulation of them (Extended Data Fig. 3c). For the other eight genes whose expression increased in Tet2-deficient dendritic cells only after LPS stimulation, Rsad2, Cmpk2 and Ifit2 had the same dynamic transcription patterns as Il6 (Extended Data Fig. 3d), and we identified binding of Tet2 to their promoters (Extended Data Fig. 3e). For lymphocytes, as shown in Extended Data Fig. 4a–c, loss of Tet2 caused higher expression of IFN-γ in activated CD8+ T cells, which have different mechanisms for transcriptional regulation of IFN-γ expression in activated CD4+ T cells and natural killer (NK) cells6. In addition, Tet2-deficient B cells expressed higher level of IL-6 but not TNF-α in response to LPS stimulation, further demonstrating the general characteristics of Tet2 in repressing IL-6 transcription (Extended Data Fig. 4d). These data indicate that Tet2 may exert its broad repression of proinflammatory genes in immune cells.
To assess the biological significance of Tet2-mediated active repression of inflammatory mediators in innate myeloid cells in vivo, we used the endotoxin shock model in myeloid cell-specific Tet2-deficient mice. The percentage of macrophages in conditional Tet2-deficient mice was not significantly affected (Extended Data Fig. 1f). Increased transcript of Il6, but not Tnf, was observed in Tet2-deficient macrophages in the late phase during LPS response (Extended Data Fig. 1j). After systemic challenge with LPS, conditional Tet2-deficient mice produced much more IL-6, especially at late phase, and their lungs exhibited more severe tissue damage and diffuse inflammation (Fig. 1d, e).
To further broaden our investigation in the physiological role of Tet2 in control of inflammatory response in vivo, we used a mouse model of colitis induced by dextran sulfate sodium (DSS) which mainly depends on the innate immune system7,8. Tet2-deficient mice were more susceptible to DSS-induced colitis and showed exacerbated colon inflammation, including exaggerated weight loss, higher production of IL-6 and infiltration of more inflammatory cells, and disruption of mucosal structures in histological analysis of the colons, as compared with control mice (Fig. 1f–h).
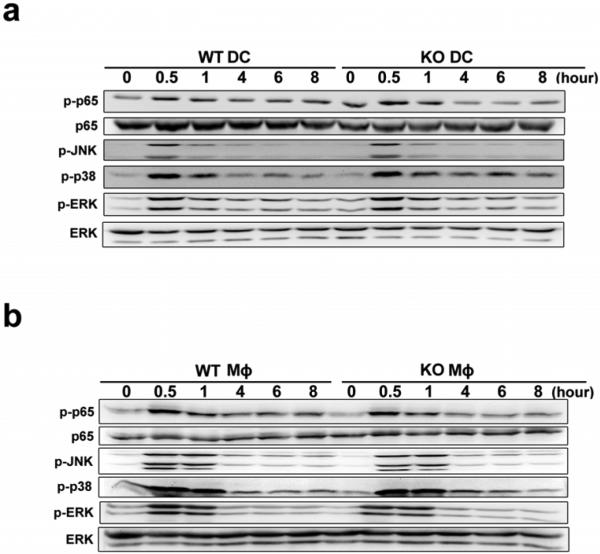
TLR signal-triggered activation of NF-κB and MAPK pathways simultaneously induce the gene expression of proinflammatory cytokines. These pathways were not affected in Tet2-deficient BMDC and peritoneal macrophages (Extended Data Fig. 5). Remarkably, although activation of these signalling pathways were equally initiated and terminated in control and Tet2-deficient cells, higher levels of IL-6 expression were maintained in Tet2-deficient cells at late phase of LPS response. Timely termination of TLR signalling pathways has been shown to control overproduction of proinflammatory cytokines and prevent inflammatory autoimmune diseases, highlighting the importance of TLR-negative regulators in inflammation resolution. However, our data indicate that termination of TLRs signalling pathways is not sufficient to attenuate transcription of Il6, and that active repression of gene transcription at chromatin level is also needed during inflammation resolution.
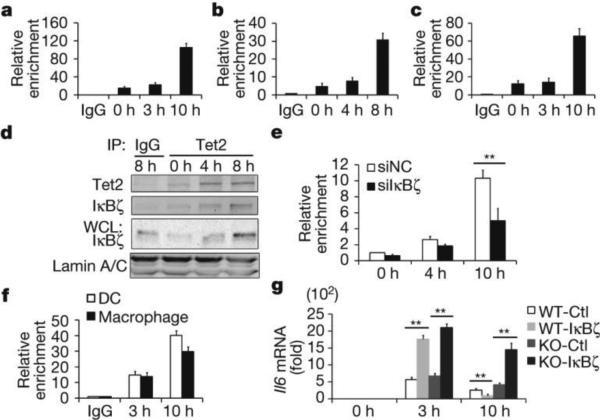
We further investigated direct targeting of Tet2 to Il6 gene, and found increased binding signal of Tet2 to the Il6 promoter at late phase during LPS stimulation (Fig. 2a–c). Unlike Tet1 and Tet3, Tet2 does not contain a CXXC DNA binding domain and may target specific genomic sites via its binding proteins, especially cell-type-specific transcription factors9. We sought functional proteins associated with endogenous Tet2 in BMDC by co-immunoprecipitation (co-IP) coupled with mass spectrometry (MS) analysis using an antibody recognizing the amino terminus of Tet2, and identified several transcription factors including IκBζ (Extended Data Fig. 6a), which has been reported to specifically target proximal Il6 promoter during LPS response10. We validated the interaction of Tet2 with IκBζ and further found that both their protein levels and their association increased at late phase during LPS response (Fig. 2d). Knockdown of IκBζ reduced the association of Tet2 with Il6 gene promoter at late phase during LPS response (Extended Data Fig. 6b and Fig. 2e). Although IκBζ enhances Il6 transcription initiation in early phase (before 4 h) during LPS response through promoting RelA binding and transcription complex assembly11, the regulatory role of IκBζ in inflammation resolution remains unclear. Accordingly, we first detected the specific binding of IκBζ at Il6 promoter, and did observe the increased association at late phase during LPS response (Fig. 2f). We further overexpressed IκBζ in primary macrophages and found that, when compared with control group, the mRNA levels of Il6 were higher at early phase but significantly lower at late phase during LPS response in IκBζ-overexpressing macrophages. Furthermore, loss of Tet2 impaired the repressive effect of IκBζ overexpression on IL-6 expression at late phase during LPS response (Fig. 2g). These results indicate that Tet2 represses Il6 transcription through directly targeting Il6 promoter, which was mediated by IκBζ at late phase during LPS response. Among the genes repressed by Tet2 in BMDC, Ccl2 and Cxcl5 expression variations differed from Il6 in BMDC at late phase during LPS stimulation (Extended Data Fig. 3c-d), although Ccl2 and Cxcl5 were also reported as targets of IκBζ10,12.
Figure 2. IκBζ mediates selective targeting of Tet2 to Il6 promoter.
a–c, e, f, ChIP followed by quantitative PCR (ChIP-qPCR) of Tet2 (a–c, e) and IκBζ (f) at the Il6 promoter in BMDC (a, f), peritoneal macrophages (b, f) and human dendritic cells (c) and IκBζ-silenced BMDC (e). IP, immunoprecipitation. d, Tet2 interacted with IκBζ in LPS-stimulated BMDC. g, Il6 mRNA in IκBζ-overexpressed wild-type and Tet2-deficient peritoneal macrophages. Cells were stimulated by LPS. Full scans of blots are shown in Supplementary Fig. 1. Error bars represent s.d. of triplicate technical replicates. Data are representative of 3 independent experiments. Unpaired Student's t-test, **P < 0.01.
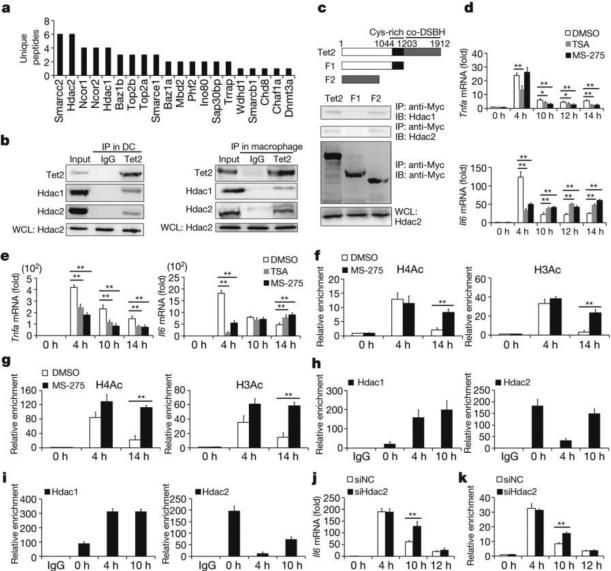
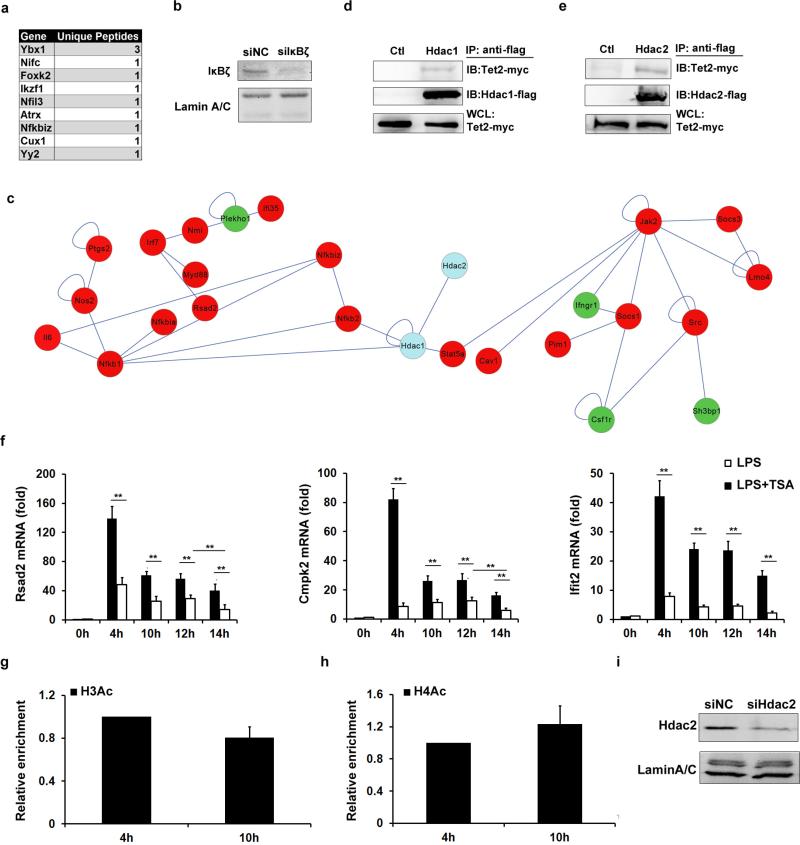
In addition to the known regulatory roles in DNA oxidation, Tet proteins, as multi-functional proteins, can interact with other epigenetic modifiers to contribute to multi-layer chromatin regulation13. Now, most of existing studies revealed that Tet2-connected O-linked glycosylation involving attachment of β-N-acetylglucosamine (GlcNAc) to Ser/Thr residues of proteins (GlcNAcylation) and H3K4 methylation to positively regulate gene transcription14,15. However, the repressive role of Tet2 has been largely unknown. We analysed functional partners for chromatin regulation of Tet2 in our mass spectrometry data, and found that Tet2 did associate with several epigenetic regulators (Fig. 3a). Surprisingly, we found that Tet2 associated with several repressive chromatin modifiers, such as members of repressive complexes Ncor1/2. Among them, two histone deacetylases, Hdac1 and Hdac2, were associated with a large number of genes with marked variations in LPS-stimulated BMDC (Extended Data Fig. 6c), indicating their potential roles in regulating innate inflammatory response together with Tet2. Their endogenous interactions in LPS-activated BMDC and peritoneal macrophages were further confirmed (Fig. 3b). To map the regions of interaction between Tet2 and Hdac1/2, we generated a series of Tet2-deletion mutants, and found that the Tet2 mutant containing the core-DSBH domain was responsible for its interaction with Hdac1/2 (Fig. 3c). In overexpression analysis, Flag-tagged Hdac1 or Hdac2 also interacted with Myc-tagged Tet2 (Extended Data Fig. 6d, e).
Figure 3. Hdac2 associates with Tet2 and specifically represses IL-6 through histone deacetylation.
a, MS-identified Tet2-binding chromatin regulators in LPS-stimulated BMDC for 8 h. b,Tet2 interacts with HDAC1/2 in cells 8 h after LPS stimulation. c, Deletion mutants of Tet2 were overexpressed to detect endogenous HDAC1/2 association. IB, immunoblot. d–i, BMDC (d, f, h) and peritoneal macrophages (e, g, i) were pre-treated with 100 nM TSA or 4 μM MS-275 (d–g), or left naive (h, i), then stimulated with LPS. Cytokine mRNA (d, e) and histone acetylation and Hdac1/2 at Il6 promoter (f–i) were analysed. j, k, Il6 mRNA and H3Ac enrichment at Il6 promoter in Hdac2-silenced (siHdac2) BMDC stimulated with LPS. Full scans of blots are shown in Supplementary Fig. 1. Error bars represent s.d. of triplicate technical (f–i, k) or s.e.m. of triplicate biological (d, e, j) replicates. Data are representative of 3 independent experiments. Unpaired Student's t-test, *P < 0.05, **P < 0.01.
Lysine acetylation on tails of histone is critical for regulating cytokine-specific transcription during innate immune response16–18. Histone deacetylase (HDAC) inhibitors can reverse silencing of several inflammatory cytokines in tolerant macrophages16. However, whether and how specifically HDACs can mediate gene-specific repression of proinflammatory cytokines during inflammation resolution have not been clearly delineated. Inhibition of HDACs significantly downregulated mRNA levels of both Tnfa and Il6 at early phase after LPS stimulation. However, the mRNA of Il6, but not Tnfa, was maintained at higher levels at late phase in the presence of HDAC inhibition (Fig. 3d, e). Although Rsad2, Cmpk2 and Ifit2 are also direct targets for Tet2-mediated repression, HDAC inhibitor did not upregulate their expression at late phase during LPS simulation, indicating the specific involvement of HDACs in repressing Il6 transcription (Extended Data Fig. 6f). We further observed dynamic patterns of H3Ac and H4Ac at the Il6 promoter that were positively correlated with transcription of IL-6, and found that MS-275 inhibited the loss of histone acetylation signals at late phase after LPS stimulation (Fig. 3f, g). MS-275 strongly inhibits Hdac1/2 over Hdac3 (ref. 19). As previously reported, Hdac3 was required for IL-6 induction by LPS in an IFN-β-dependent mechanism20. Thus, we investigated the roles of Hdac1/2 for Il6 transcription repression. ChIP analysis showed that the binding of Hdac1 increased and the binding of Hdac2 decreased to the Il6 promoter in early phase during LPS stimulation, while the binding of Hdac2 was restored at late phase (Fig. 3h, i). For TNF-α, we did not observe significant histone deacetylation at its promoter in late time phase during LPS response (Extended Data Fig. 6g, h). We further found higher transcription and histone acetylation levels at the Il6 gene locus in Hdac2-silenced dendritic cells at 10 h after LPS stimulation (Fig. 3j, k and Extended Data Fig. 6i). However, significant compensation for Hdac2 loss, probably mediated by Hdac1, led to similar transcription and histone acetylation levels with the control groups at 12 h after LPS stimulation in Hdac2-silenced dendritic cells. These results indicated that Hdac2 was the key enzyme in histone deacetylation-mediated inhibition of IL-6 during inflammation resolution.
To further reveal the in vivo recruitment of Hdac2 to Il6 promoter by Tet2, we performed sequential ChIP (re-ChIP) and found that Tet2 and Hdac2 formed a complex to bind Il6 promoter in LPS-activated dendritic cells. Considering that Hdac1 may compensate for the loss of Hdac2 in Il6 locus, we also detected association of Tet2 with Hdac1, and found that Hdac1 was indeed in the complex (Fig. 4a). Furthermore, using the principle of yeast two hybrid system, we found that Tet2 fused with the GAL4 DNA binding domain was sufficient to recruit both Hdac2 and Hdac1 to the GAL4 DNA binding sites in macrophages (Fig. 4b).
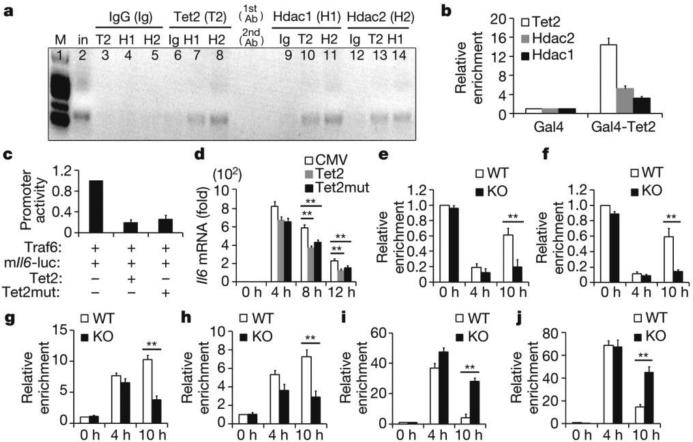
Figure 4. Tet2 recruits Hdac2 to repress expression of IL-6.
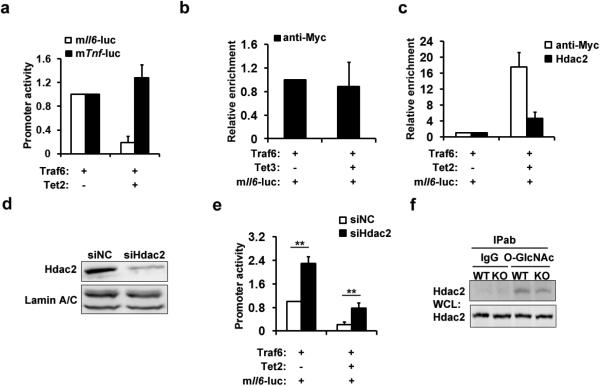
a, Re-ChIP-PCR assay of Tet2–Hdac1/2 interaction at Il6 promoter in BMDC 8 h after LPS stimulation. First round ChIP (1st antibody, Ab) against Tet2 (T2), Hdac1 (H1), Hdac2 (H2) or IgG (Ig). Eluted samples were re-ChIPed (2nd antibody). Lane 2 was input (in). b, ChIP-qPCR assay of Tet2 and Hdac1/2 at Gal4-binding sites in Gal4-Tet2-overexpressing peritoneal macrophages. c, Luciferase activities in lysates of HEK293T cells transfected with indicated plasmids and murine Il6 promoter reporter (mIl6-luc). d, Il6 mRNA in wild-type Tet2- or catalytic mutant Tet2-overexpressed Tet2-deficient peritoneal macrophages stimulated with LPS. e–j, Tet2-deficient BMDC (e, g, i) and peritoneal macrophages (f, h, j) were stimulated with LPS. Hdac2 (e, f), Hdac1(g, h), H3Ac (i), H4Ac (j) enrichments at Il6 promoter were analysed by ChIP-qPCR. Full scan of gel is shown in Supplementary Fig. 1. Error bars represent s.d. of triplicate technical replicates (b, d–j) or s.e.m. of triplicate biological replicates (c). Data are representative of 3 independent experiments. Unpaired Student's t-test, **P < 0.01.
Overexpression of Tet2 repressed TLR signal-induced luciferase activity of Il6 promoter but not of the Tnf promoter (Extended Data Fig. 7a). We did not detect binding of overexpressed Tet3 to the Il6 promoter (Extended Data Fig. 7b). Compared with wild-type Tet2, an enzymatic mutation form of Tet2 had a similar repressive effect on both luciferase Il6 promoter activity and Il6 transcription (Fig. 4c, d). Furthermore, overexpressed Tet2 recruited more endogenous Hdac2 to the Il6 promoter (Extended Data Fig. 7c). Silencing of Hdac2 not only increased the TLR signal-induced activation of Il6 promoter in control group, but also attenuated the repressive effect of Tet2 over-expression (Extended Data Fig. 7d, e).
We further confirmed the recruitment of Hdac2 by Tet2 to Il6 promoter using Tet2-deficient dendritic cells and macrophages, and found that loss of Tet2 significantly decreased the binding of Hdac2 to Il6 promoter (Fig. 4e, f), and association of Hdac1 was also significantly decreased at late phase during LPS response owing to Tet2 loss (Fig. 4g, h). Attenuated histone deacetylation in Tet2-deficient cells was also observed at late phase during LPS response (Fig. 4i, j). Although Tet2 interacts with O-GlcNAc transferase (OGT) and facilitates OGT-dependent O-GlcNAcylation14,15, loss of Tet2 did not affect O-GlcNAcylation of Hdac2 (Extended Data Fig. 7f). Furthermore, our results also indicate that Hdac1 is in the complex containing Tet2 and Hdac2, and Tet2 and Hdac2 can stabilize association of Hdac1 with the Il6 promoter, especially during inflammation resolution. Together, these results demonstrate that Tet2 mediates active repression of IL-6 through HDACs to erase histone acetylation during inflammation resolution. Furthermore, intergenic regions with low CpG densities, like the promoter of Il6, may involve Tet proteins in transcription regulation. Conversely the Tnf promoter contains CpG islands, in which DNA methylation has essential roles in transcription regulation via other chromatin modifiers.
Here, our data further indicate that epigenetic modifier-mediated active chromatin regulation in a gene-specific manner is a critical factor in repressing inflammatory gene transcription during inflammation resolution. Beside IL-6, we have also identified several other well-known proinflammatory mediators which contribute to tissue damage and inflammatory cell infiltration during inflammation response and are repressed by Tet2. Furthermore, considering that epigenetic regulation of gene expression is effected in a cell-specific manner, there may be more proinflammatory mediators repressed by Tet2 in tissue resident immune cells, which also contributes to proinflammatory phenotype due to Tet2 loss.
Dysfunction of Tet2 has a known role in the pathogenesis of myeloid malignancies21,22. Here we describe a novel role for Tet2 in regulating inflammation, which may also provide clues to investigate how Tet2 contributes to the pathogenesis of myeloid cancer, beyond the intrinsic role of Tet2 in leukaemia cells themselves. Moreover, IL-6 is now regarded as a prominent target for clinical intervention in the treatment of inflammation, autoimmunity and cancer23,24. Our data suggest that targeting Tet2/Hdac2-mediated gene-specific repression may represent a novel therapeutic approach in an epigenetic view.
METHODS
Mice and reagents
C57BL/6 mice were obtained from Joint Ventures Sipper BK Experimental Animal Company (Shanghai, China) and used at the age of about 6 weeks. Tet2fl/fl mice and Tet2-deficient mice on a C57BL/6 × 129/SvEv background were kindly provided by R.L.L. Lyz2-Cre mice were from The Jackson Laboratory. All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Second Military Medical University (SMMU), Shanghai. LPS, TSA and anti-Flag (M2) were from Sigma-Aldrich. Recombinant GM-CSF and IL-4 were from R&D Systems. Tet2 (MABE462), H3Ac (06-599) and H4Ac (06-866) antibodies were from Millipore. TET3 (C3) antibody was from GeneTex. Hdac1 (C-19), Hdac2 (C-19), p65 (C-20), ERK (H-72), JNK (FL), p38 (C-20)and LaminA/C (H-110) antibodies and MS-275 were from Santa Cruz. IκBζ antibody (A311-451A) was from Bethyl Labs. RL2 monoclonal antibody against O-GlcNAc was from Abcam. Antibody to Myc-Tag (9B11), p65 phosphorylated at Ser536 (3031), ERK phosphorylated at Thr202/Tyr204 (20G11), JNK phosphorylated at Thr183/Tyr185 (81E11) and p38 phosphorylated at Thr180/Tyr182 (28B10) were from Cell Signaling Technology. Magnetic beads coated with antibodies for cell isolation were from Miltenyi Biotec and cell marker antibodies were purchased from BD Biosciences22,25.
Cell purification and cultures
Using Ficoll-paquePLUS (GE healthcare), human peripheral blood mononuclear cells (PBMNC) were freshly isolated from peripheral blood samples of healthy donors. The study was approved by the ethics committees of the Second Military Medical University (SMMU), Shanghai and informed donor consent was obtained. The PBMNC were stained with anti-CD14 mAb (BD), and the monocytes (purity >99%) were sorted using a MoFloXDP High-performance Cell Sorter (DACO Cytomatix). The monocytes were cultured in RPMI 1640 (GIBCO) supplemented with 10% heat-inactivated FBS (Invitrogen), GM-CSF (25 ng ml−1) and IL-4 (10 ng ml−1). Thioglycollate-elicited murine peritoneal macrophages (peritoneal macrophages), BMDC and macrophages (BMM) (20 ng ml−1 M-CSF) were prepared as previously reported26. 100 ng ml−1 LPS was used for stimulation. Cells were cultured in endotoxin-free DMEM (GIBCO) supplemented with 10% (v/v) FBS. Preparation of splenic NK cells, CD4+T cells, CD8+T cells and B cells were conducted with purification system according to the manufacturer's protocol as described before25. HEK293T cells were from ATCC and cultured in endotoxin-free RPMI1640 supplemented with 10% (v/v) FBS without further authentication and mycoplasma contamination test.
RNA-seq analysis
Total RNA was used for mRNA isolation and cDNA library generation with the TruSeq RNA Sample Preparation Kit (Illumina). Clusters were generated with the TruSeq SR Cluster Kit v2 according to the reagent preparation guide. The RNA sequencing was performed using the Illumina platform. High-quality reads were aligned to the mouse reference genome (mm9) using SOAP aligner. The expression levels for each of the genes were normalized to reads per kilobase of exon model per million mapped reads (RPKM) to compared mRNA levels between samples.
Lentivirus preparation
Lentivirus were prepared and infected as previously described25. For Tet2 inhibition, the sequence encoding short interfering RNA (siRNA) for Tet2 (5′-CCAUCACAAUUGCUUCUUU-3′) was cloned into a pSIF-H1-copGFP vector (catalogue no. SI101B-1, System Bioscience).
ChIP and re-ChIP assay
The cells were cross-linked with 1% (v/v) methanol-free formaldehyde for 10 min (histone modification) or 20 min (chromatin modifiers) and processed according to the protocol described in Chromatin Immunoprecipitation (ChIP) Assay Kit (Millipore). Re-ChIP assays were performed using Re-ChIP-IT kit (Active motif) with specific antibodies. Antibody-chromatin complexes were pulled-down using magnetic protein G beads (Cell Signaling Technology), washed and then eluted. After cross-link reversal and proteinase K treatment, immunoprecipitated DNA was extracted with phenol-chloroform, ethanol precipitated. ChIP primers were as follows:
mIL-6pro(0-0.5K)-F: cctgcgtttaaataacatcagctttagctt, R: gcacaatgtgacgtcgtttagcatcgaa; mTNFpro(0-0.5K)-F: ccagccagcagaagctccctcagcgag, R: gcggatcatgcttt ctgtgctca tggtgtc; hIL-6pro(0-0.5K)-F: acttcgtgcatgacttcagc, R: agtgcagcttaggtcgtcat; mRsad2pro(0-0.5K)-F: tcttggctctggtccaactt, R: actgtgtcacaaggaggagg; mCmpk 2pro(0-0.5K)-F: ggaattctcaagagcaggcg, R: taggaaattctggccctggg; mIfit2pro (0-0.5K)-F: accgtctctctcccaattcc, R: ctgtgtcctgctattgtcgc.
qPCR assay
For mRNA analysis, LightCycler (Roche) and SYBR RT–PCR kit (TOYOBO) were used for qPCR analysis as previously described27. The 2−ΔΔCt change-in-cycling-threshold method was used for calculation of relative changes in expression. Signals were normalized by β-actin, and compared with control groups. For quantification in ChIP assay, primers with the length of their amplicons between 50-250bp for ChIP-qPCR were designed using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0) and in-silico validated in UCSC genome browser for specificity. The efficiency of these primers was evaluated by qPCR with gradually diluted template DNA. Dissociation plot and DNA agarose electrophoresis of amplicons were evaluated for qPCR specificity. Signals were normalized and calculated using 1% input and used for comparison between experimental samples. When antibody against target protein was initially used in one type of cells, normalized signals in IgG groups were calculated and used for comparison with experimental samples. When normalized signals in experimental sample is at least 4 times as high as that in IgG group, the enrichment were considered positive for subsequent experiments. For controls used for comparison, if IgG group was contained, all experiment samples were compared with it, or else, experimental sample indicated as control in figure legend is compared with the others.
Transfection
Cells were seeded and maintained overnight, then transfected with gene-specific siRNAs via INTERFERin (Polyplus) for 36–48 h, and stimulated with LPS (100 ng ml−1). Sequences of siRNAs were as follows:
Tet2: 5′-GGAAUAUCCCACAUGAAAGGCAGCC-3′
Tet3: 5′-CCCACAAGGACCAACAUAA-3′
Hdac2:5′-CCCAAUGAGUUGCCAUAUA-3′
IκBζ:5′-GCUGCCCUGCUUCAGAAUA-3′
For overexpression assay in primary cells, thioglycollate-elicited murine peritoneal macrophages were transfected by nucleofection with a Mouse Macrophage Nucleofector kit (Amaxa)28.
Plasmid constructs
Full-length cDNAs of murine Tet2, Tet3 or IκBζ were PCR amplified using cDNA from peritoneal macrophages. Tet2 fragments were amplified according to previously reported domains29. Enzymatic activity of Tet2 was mutated (H1304Y and D1306A) as previously reported5,29. Tet2, Tet2-derived and Tet3 amplicons were subcloned into the pCMV-Myc-N (Clontech) or pBIND (Promega) vectors. IκBζ was subcloned into the pcDNA3.1 (Invitrogen) vectors. All constructs were confirmed by DNA sequencing. Luciferase reporter plasmids for Tnf and Il6 promoters (TSS to upstream 3K) were constructed into pGL3 luciferase reporter vectors (Promega). Gal4 binding site containing reporter (GAL4/UAS system) used in Gal4-Tet2 recruitment assay was from Promega.
Immunoprecipitation (IP) and immunoblot (IB) analysis
These assays were performed as reported24. Cells were lysed with Cell Lysis Buffer or RIPA buffer (Cell Signaling Technology) with supplemented with a protease-inhibitor ‘cocktail’ (Roche). Protein concentrations in the extracts were measured by BCA assay. The whole-cell lysates (WCL) were used as input and loading control.
Co-immunoprecipitation coupled with LC–MS/MS analysis
108 murine BMDC stimulated with LPS for 8 h were swelled 20 min in 50 ml RSB buffer (10 mM Tris-HCl pH 8.0, 10 mM NaCl, 0.1% NP40 (v/v), 3 mM MgCl2), and centrifuged at 2,000g for 10 min at 4 °C. Pellet containing the nucleus were lysed with 10 ml Cell Lysis Buffer and sonicated to break the nucleus and fragment the chromosome. Soluble fraction was subjected to immunoprecipitation with 10 μg antibody recognizing N terminus of Tet2 overnight. 100 μl protein G Magnetic beads (Cell Signaling Technology) prewashed with Cell Lysis Buffer containing 0.1% BSA were added and incubated for another 2 h at 4 °C. Protein complex-containing beads were washed with NETN (300 mM NaCl, 20 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.05% (v/v) NP-40) four times. Proteins were eluted and boiled in 1% SDS loading buffers. After SDS–PAGE and silver staining, positions in gel lanes where there were more bands in Tet2-group were determined. Gel pieces at these positions in both IgG- and Tet2-group were respectively cut and subjected into in-gel digestion and LC–MS/MS analysis. The resulting MS/MS data were processed using Mascot search engine (version 3.2). Tandem mass spectra were searched against SwissProt-Mouse database concatenated with reverse decoy database. Proteins containing unique mapped peptide with ion score meeting significant P value were considered. These eligible proteins appeared in Tet2-group but not in IgG-group were chosen.
Assay of luciferase reporter gene expression
HEK293T cells were transfected with a mixture of the appropriate luciferase reporter plasmid, pRL-TK-renillaluciferase plasmid, Traf6 expression plasmid and the appropriate additional constructs for 36 h using jetPEI (Polyplus). The total amount of plasmid DNA was equalized by empty control vector. Alternatively, HEK293T cells were transfected with plasmids as above plus Hdac2 siRNA (5′-CCAUGAAGCCUCAUAGAAU-3′) for 48 h using jetPRIME (Polyplus). Luciferase activity was measured with a Dual-Luciferase Reporter Assay System according to the manufacture's protocols (Promega) after 36 h. Data were normalized for transfection efficiency by the division of firefly luciferase activity with that of Renilla luciferase.
Statistical analysis
Error bars displayed throughout the manuscript represent s.e.m. or s.d. and were calculated from triplicate technical or triplicate biological replicates described in figure legends. Sample sizes were chosen by standard methods to ensure adequate power, and no randomization of weight and sex or blinding was used for animal studies. Data shown are representative of 3 independent experiments, including histological images, blots and gels. No statistical method was used to predetermine sample size. Statistical significance was determined using unpaired Student's t-tests; *P < 0.05; **P < 0.01.
Extended Data
Extended Data Figure 1. Loss of Tet2 specifically enhances expression of IL-6 but not TNF-α.
a, Hierarchical cluster of transcription level of chromatin modifiers with significant variation in LPS-stimulated murine BMDC, RPKMs were centred by the mean. b,Tet2 mRNA in human dendritic cells (hDC), LPS-stimulated murine BMDC, BMM and primary peritoneal macrophages (pMϕ). c–f, j, Human imDCs in which TET2 were lentivirally silenced (LV-siTET2), BMDC, BMM and peritoneal macrophages from Tet2-deficient mice (c–f), peritoneal macrophages from conditional Tet2-deficient mice (f, j) and their respective controls were stimulated by LPS for 8 h (c) or the indicated time (d, e, j). Protein levels of TET2 were detected by immunoblot, with lamin A/C as the loading control (c). IL-6 protein levels were analysed by ELISA (d), and Tnfa and Il6 mRNA levels were analysed by qPCR (e, j). Percentage of Iab+CD11c+BMDC or F4/80+ CD11b+peritoneal macrophages were analysed (f). g–i, Tet2 (g, h), Tet3 (g, i) were silenced in peritoneal macrophages for 48 h and then peritoneal macrophages were stimulated with LPS for 8 h (g, i) or the indicated time (h), and the protein levels of Tet2 or Tet3 were detected by immunoblot, with lamin A/C as the loading control (g). Tnfa and Il6 mRNA were analysed by qPCR (h, i). Full scans of blots are shown in Supplementary Fig. 1. Error bars represent s.e.m. of triplicate biological replicates and are representative of three independent experiments. **P < 0.01.
Extended Data Figure 2. Loss of Tet2 barely affects development of immune cells.
Percentages of the indicated immune cells were analysed by their specific lineage markers from control and Tet2-deficient mice. Data were from one representative of three independent experiments.
Extended Data Figure 3. Gene expression variation in Tet2-deficient dendritic cells.
a, b, mRNA variations of indicated genes in RNA-seq analysis of BMDC stimulated with LPS. RPKM of each of the genes in 4 h group were compared with 0 h group, and calculated to log2 ratio. c, d, qPCR analysis of mRNA levels of indicated genes in wild type and Tet2-deficient naive BMDC (c) or LPS-stimulated BMDC (d). e, BMDC were stimulated with LPS for 8 h, and then endogenous Tet2 enrichments at proximal promoter of indicated genes were analysed by ChIP-qPCR. Error bars represent s.d. of triplicate technical (e) or s.e.m. of triplicate biological (c, d) replicates and are representative of three independent experiments. **P < 0.01.
Extended Data Figure 4. Potential targets of Tet2 during immune response.
a–d, ELISA of cytokines in supernatants of splenic activated CD4+ T cells (a), CD8+ T cells (b) and NK cells (c), and conventional splenic CD19+ B cells (d) from Tet2-deficient (KO) and control mice (WT). T cells were stimulated with 10 μg ml−1 anti-CD3 and 1 μg ml−1 anti-CD28 monoclonal antibodies for 24 h. NK cells were stimulated with a combination of 10 ng ml−1 IL-12 plus 5 ng ml−1 IL-18 for 12 h. B cells were stimulated with 5 μg ml−1 LPS for 24 h, Error bars represent s.d. of triplicate technical replicates and are representative of three independent experiments. **P < 0.01.
Extended Data Figure 5. Loss of Tet2 barely affects TLR4 signalling pathways.
a, b, Immunoblot assays of the phosphorylated (p-) or total proteins in lysates of wild-type and Tet2-deficent BMDC (a) and peritoneal macrophages (b) stimulated with LPS for indicated time. Full scans of blots are shown in Supplementary Fig. 1. Data were from one representative of three independent experiments.
Extended Data Figure 6. Tet2 binding partners for cytokine regulation.
a, Co-immunoprecipitation assay using Tet2 antibody of nuclear fraction of murine BMDC stimulated with LPS for 8 h. Gene symbols of transcription factor deposited in KEGG database and their unique peptides identified by mass spectrometry analysis were tabled. b, IκBζ was silenced in BMDC using specific siRNA for 36 h and the BMDC were stimulated with LPS for 8 h. The protein levels of IκBζ were detected by immunoblot, with lamin A/C as the loading control. c, Hdac1/2 (blue circles) were subjected to interaction analysis with genes which had significant expression variations (red circles for upregulated, and green circles for downregulated) in BMDC 4 h after LPS stimulation. The linkers indicate interaction or regulation relationship between the two genes. d, e, Flag-tagged Hdac1 (d) or Hdac2 (e) were overexpressed in HEK293T cells together with Myc-tagged Tet2. Cell lysates were examined by IP and immunoblot with indicated antibodies. The whole-cell lysates (WCL) were used to examine the input of overexpressed proteins. f, BMDC were pretreated with 100 nM TSA for 1 h, and then stimulated by LPS for indicated time. mRNA levels of indicated genes were analysed by qPCR. g, h, BMDC (g) and peritoneal macrophages (h) were stimulated with LPS for the indicated time. Enrichments of H3Ac (g), H4Ac (h) at Tnf promoters were analysed by ChIP-qPCR. i, Hdac2 was silenced in BMDC for 48 h and then the BMDC were stimulated with LPS for 8 h. The protein levels of Hdac2 were detected by immunoblot, with lamin A/C as the loading control. Full scans of blots are shown in Supplementary Fig. 1. Error bars represent s.d. of triplicate technical (g, h) or s.e.m. of triplicate biological (f) replicates and are representative of three independent experiments. **P < 0.01.
Extended Data Figure 7. Tet2 represses Il6 promoter activity through recruiting Hdac2.
a, Luciferase activities in lysates of HEK293T cells transfected with indicated plasmids and luciferase reporter for murine Il6 promoter (TSS to upstream 3K). b, ChIP-qPCR analysis of Myc-tagged Tet3 in murine Il6 promoter in HEK293T cells which were transfected with indicated plasmids and luciferase reporter. c, ChIP-qPCR of Myc-tagged Tet2 or endogenous Hdac2 in murine Il6 promoter in HEK293T cells transfected with indicated plasmids and luciferase reporter. d, Hdac2 was silenced for 36 h in HEK293T cells. Protein levels of Hdac2 were detected by immunoblot, with lamin A/C as the loading control. e, Luciferase activities in lysates of Hdac2-silenced HEK293T cells transfected with indicated plasmids and luciferase reporter containing murine Il6 promoter. f, Wild-type (WT) and Tet2-deficient (KO) peritoneal macrophages were stimulated with LPS for 8 h. IP using O-GlcNAcylation antibody was performed. The whole-cell lysates (WCL) were used as the input. Full scans of blots are shown in Supplementary Fig. 1. Error bars represent s.d. of triplicate technical replicates and are representative of three independent experiments. **P < 0.01.
Supplementary Material
Acknowledgements
This work was supported by the National Key Basic Research Program of China (2013CB530503) and the National Natural Science Foundation of China (31200654, 31390431, 81230074, 81123006).
Footnotes
Author Contributions Q.Z., K.Z., Q.S. and Y.H. performed the experiments, analysed the data and contributed equally for the whole study. Y.G., X.L., D.Z., Y.L., C.W., X.Z., X.S., J.L., W.G. and N.L. provided reagents and performed experiments; RL.L. provided Tet2 KO mice. X.C. and Q.Z. analysed the data and wrote the manuscript. X.C. designed and supervised the study.
Author Information The deep sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE69256.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
References
- 1.Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nature Rev. Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 7.Tlaskalová-Hogenová H, et al. Involvement of innate immunity in the development of inflammatory and autoimmune diseases. Ann. NY Acad. Sci. 2005;1051:787–798. doi: 10.1196/annals.1361.122. [DOI] [PubMed] [Google Scholar]
- 8.Neurath MF. Cytokines in inflammatory bowel disease. Nature Rev. Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 9.Ichiyama K, et al. The methylcytosine dioxygenase tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto M, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 11.Kayama H, et al. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IκBζ. J. Biol. Chem. 2008;283:12468–12477. doi: 10.1074/jbc.M709965200. [DOI] [PubMed] [Google Scholar]
- 12.Hildebrand DG, et al. IκBζ is a transcriptional key regulator of CCL2/MCP-1. J. Immunol. 2013;190:4812–4820. doi: 10.4049/jimmunol.1300089. [DOI] [PubMed] [Google Scholar]
- 13.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deplus R, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 17.Qiao Y, et al. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue S, Mai A, Dyer MJ, Cohen GM. Inhibition of histone deacetylase class I but not class II is critical for the sensitization of leukemic cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2006;66:6785–6792. doi: 10.1158/0008-5472.CAN-05-4563. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl Acad. Sci. USA. 2012;109:E2865–E2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rampal R, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, et al. Rhbdd3 controls autoimmunity by suppressing the production of IL-6 by dendritic cells via K27-linked ubiquitination of the regulator NEMO. Nature Immunol. 2014;15:612–622. doi: 10.1038/ni.2898. [DOI] [PubMed] [Google Scholar]
- 24.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nature Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 25.Ma F, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nature Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Histone lysine methyltransferase Ezh1 promotes TLR-triggered inflammatory cytokine production by suppressing Tollip. J. Immunol. 2015;194:2838–2846. doi: 10.4049/jimmunol.1402087. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013;152:467–478. doi: 10.1016/j.cell.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Xia M, et al. Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity. 2013;39:470–481. doi: 10.1016/j.immuni.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.