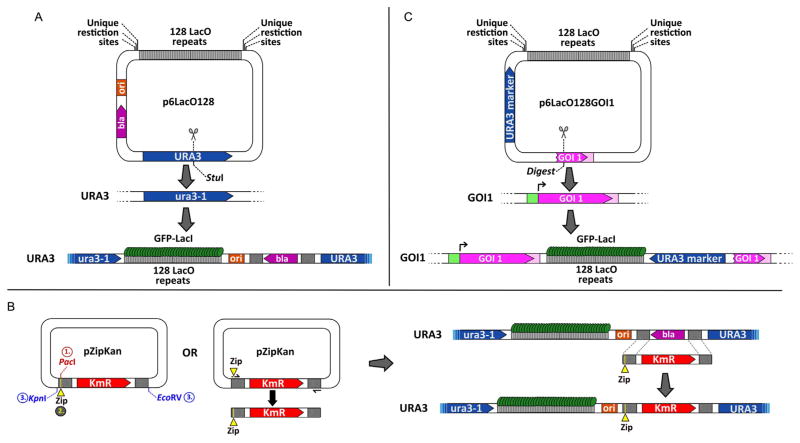
Figure 21.1. Methodology Used in Strain Construction for Microscopy.
Inserting the LacO array and testing sequences for zip code activity in the yeast genome. (A) The p6LacO128 plasmid is integrated at URA3 by digestion with StuI. Putative DNA zip codes can be cloned bedside the LacO array using the unique restriction sites on either side of the array. (B) Integration of putative zip codes (yellow Zip segments) next to the LacO array already incorporated within yeast genome through Kanr/Ampr exchange. Left: Putative DNA zip codes are either cloned into the PacI site in the pZipKan plasmid and then introduced into yeast by digestion with KpnI+EcoRV or incorporated into the cassette by including the putative zip code in the primer and amplifying the cassette by PCR. Right: The KmR cassette with a zip code can be transformed into a yeast strain having the LacO array integrated at URA3. Transformants are selected on G418 medium and screened by PCR for proper integration. (C) General strategy to incorporate the LacO array next to a gene of interest (GOI1). The 3′-end and 3′-UTR of the gene of interest is cloned into p6LacO128. Digestion within this sequence at a unique site promotes integration of the LacO plasmid downstream of the gene of interest. Ura+ transformants are selected on medium lacking uracil.