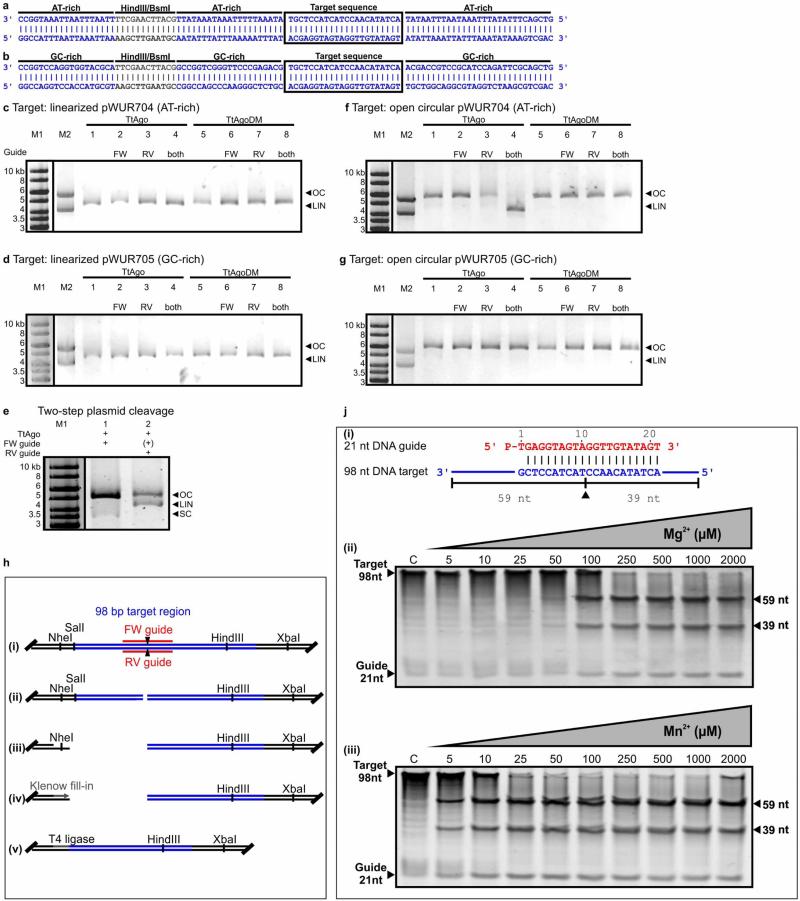
Extended Data Figure 5. Activity analyses of TtAgo.
a, b, AT-rich (17% GC) insert of pWUR704 (a) and GC-rich insert (59% GC) of pWUR705 (b). The target sequence is boxed. Restriction sites HindIII and BsmI are indicated in grey. Sequences are displayed in the 3′–5′ direction to allow comparison with Fig. 4b, which shows guide base pairing to this sequence. c, d, SpeI-linearized plasmid pWUR704 (c) and pWUR705 (d) incubated with TtAgo–siDNA and TtAgoDM–siDNA complexes targeting both strands of the plasmid, and resolved on 0.8% agarose gels. LIN, linear; M1, 1 kb Generuler marker (Fermentas); M2, open circular and linearized pWUR704 (c), or open circular and linearized pWUR705 (d); OC, open circular. FW guide: BG3466. RV guide: BG4017. High salt concentration (250 mM NaCl) in the reaction buffer cause the TtAgo-treated samples to run higher in the gel than M1 and M2. e, Two-step plasmid cleavage. Target pWUR704 was first nicked by a TtAgo–siDNA complex targeting the first strand (FW guide, lane 1), after which a TtAgo–siDNA complex targeting the other strand was added (RV guide, lane 2). FW guide is still present, and its presence is therefore indicated as (+). LIN, linear; M1, 1 kb Generuler marker (Fermentas); OC, open circular; SC, supercoiled. f, g, Nb.BsmI-nicked plasmid pWUR704 (f) and pWUR705 (g) incubated with TtAgo–siDNA and TtAgoDM–siDNA complexes targeting the un-nicked strands of the plasmid (33 bp away from the nicking site), and resolved on 0.8% agarose gels. LIN, linear; M1, 1 kb Generuler marker (Fermentas); M2, open circular and linearized pWUR704 (a), or open circular and linearized pWUR705 (b); OC, open circular. High salt concentrations (250 mM NaCl) in the reaction buffer cause the TtAgo-treated samples to run higher in the gel than M1 and M2. h, TtAgo dsDNA cleavage site analysis. (i) Plasmid pWUR704 with TtAgo–siDNA target sequences. Predicted cleavage sites are indicated with black triangles. (ii) pWUR704 was linearized using TtAgo–siDNA complexes targeting the plasmid on both strands. (iii) The linearized plasmid was cleaved using either NheI (as shown) or XbaI (not shown). (iv) Restriction site overhangs and possible overhangs resulting from TtAgo–siDNA cleavage were filled using Klenow fragment polymerase (Fermentas). (v) Blunt-end DNA was ligated using T4 DNA ligase (Fermentas), after which the plasmid could be transformed and later sequenced to determine the cleavage site. Sequences revealed that TtAgo–siDNA complexes indeed cleaved the target at the predicted locations (as shown in a), and are shown in more detail in Fig. 4b and Extended Data Fig. 5a, b. Note that in this picture target sequences are displayed in reversed order compared with Fig. 4b and Extended Data Fig. 5a, b. j, TtAgo prefers Mn2+ over Mg2+ as a divalent cation for cleavage. (i) 21-nucleotide DNA guide and 98-nucleotide ssDNA target used. The predicted cleavage site is indicated with a black triangle. (ii) 98-nucleotide ssDNA target cleavage reaction with TtAgo loaded with a 21-nucleotide siDNA in the presence of increasing concentrations of Mg2+, as indicated on top of the gel. (iii) 98-nucleotide ssDNA target cleavage reaction with TtAgo loaded with a 21-nucleotide siDNA in the presence of increasing concentrations of Mn2+, as indicated. Samples were resolved on 15% denaturing polyacrylamide gels.