Abstract
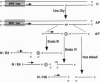
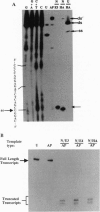
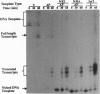
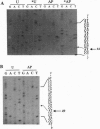
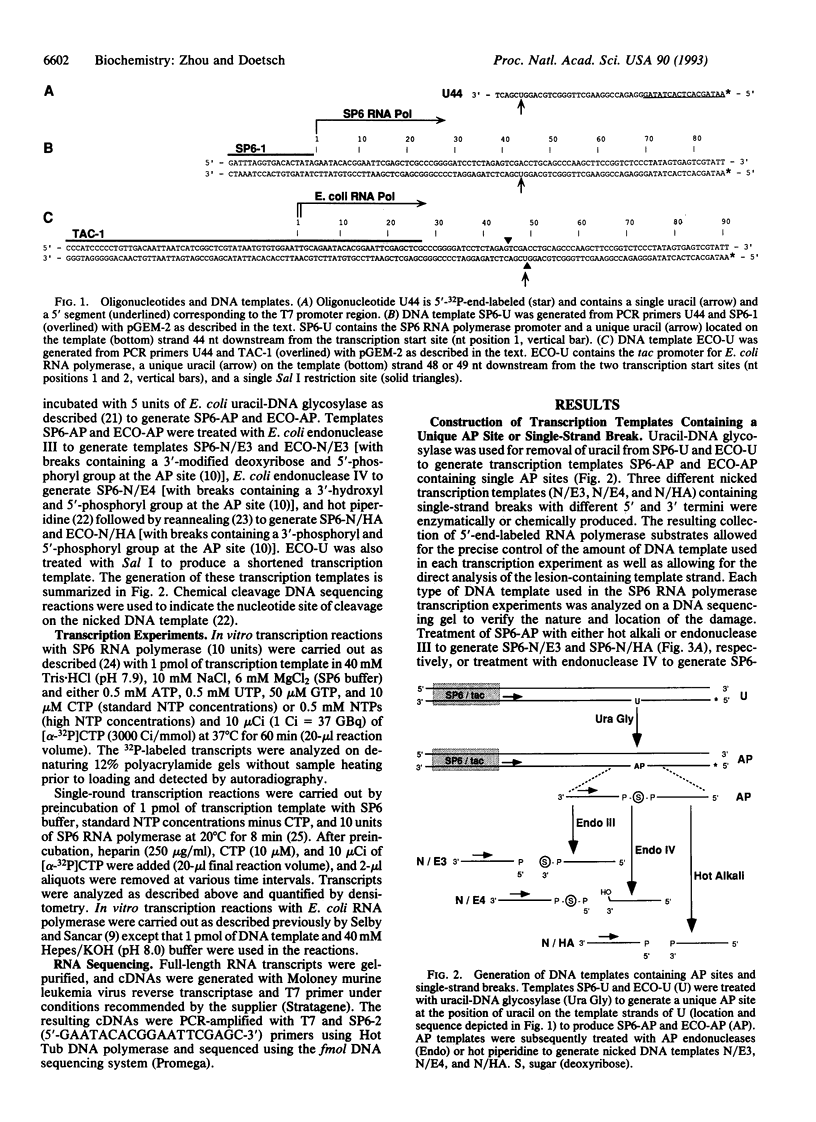
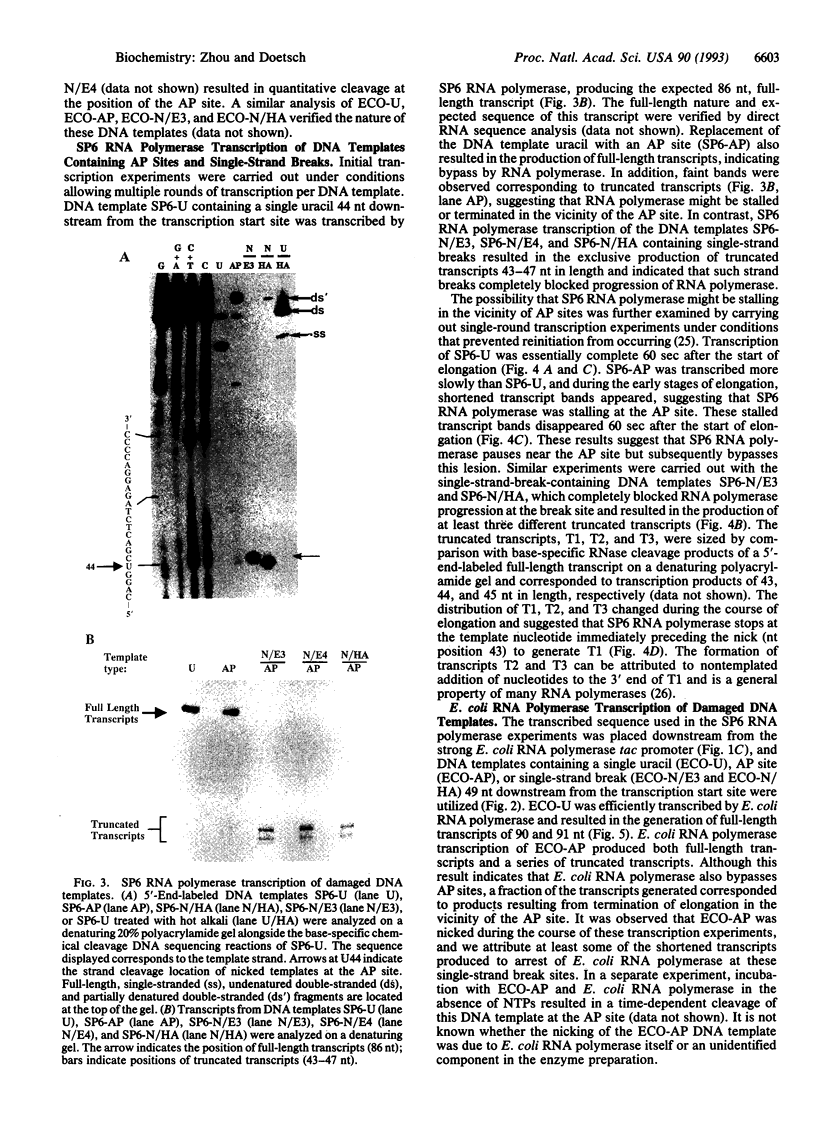
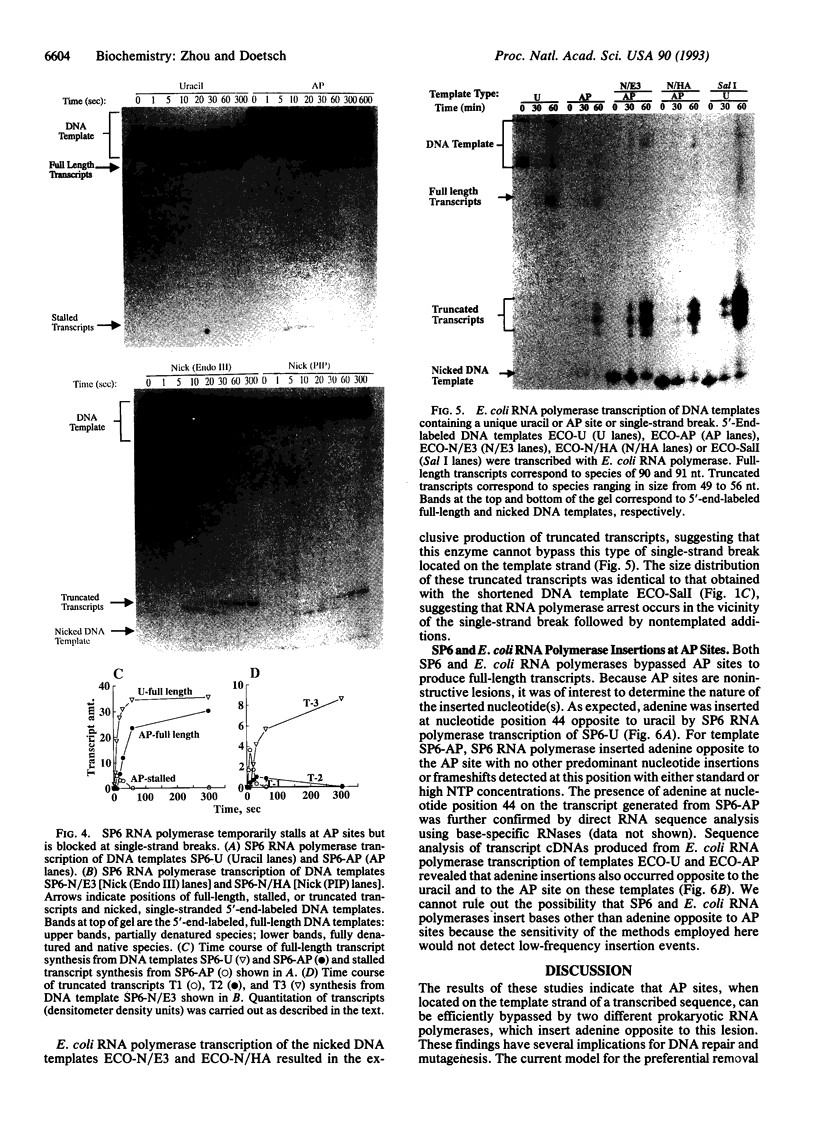
Abasic sites are thought to be the most frequently occurring cellular DNA damage and are generated spontaneously or as the result of chemical or radiation damage to DNA. In contrast to the wealth of information that exists on the effects of abasic sites on DNA polymerases, very little is known about how these lesions interact with RNA polymerases. An in vitro transcription system was used to determine the effects of abasic sites and single-strand breaks on transcriptional elongation. DNA templates were constructed containing single abasic sites or nicks placed at unique locations downstream from two different promoters and were transcribed by SP6 and Escherichia coli RNA polymerases. SP6 RNA polymerase is initially stalled at abasic sites with subsequent, efficient bypass of these lesions. E. coli RNA polymerase also bypassed abasic sites. In contrast, single-strand breaks introduced at abasic sites completely blocked the progression of both RNA polymerases. Sequence analysis of full-length transcripts revealed that SP6 and E. coli RNA polymerases insert primarily, if not exclusively, adenine residues opposite to abasic sites. This finding suggests that abasic sites may be highly mutagenic in vivo at the level of transcription.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boiteux S., Laval J. Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry. 1982 Dec 21;21(26):6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- Butler E. T., Chamberlin M. J. Bacteriophage SP6-specific RNA polymerase. I. Isolation and characterization of the enzyme. J Biol Chem. 1982 May 25;257(10):5772–5778. [PubMed] [Google Scholar]
- Chen Y. H., Matsumoto Y., Shibutani S., Bogenhagen D. F. Acetylaminofluorene and aminofluorene adducts inhibit in vitro transcription of a Xenopus 5S RNA gene only when located on the coding strand. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9583–9587. doi: 10.1073/pnas.88.21.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane C., Phillips D. R. In vitro transcription analysis of DNA adducts induced by cyanomorpholinoadriamycin. Biochemistry. 1992 Oct 13;31(40):9513–9519. doi: 10.1021/bi00155a001. [DOI] [PubMed] [Google Scholar]
- Doetsch P. W., Cunningham R. P. The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res. 1990 Sep-Nov;236(2-3):173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- Flamée P. A., Verly W. G. Action of intact AP (apurinic/apyrimidinic) sites and AP sites associated with breaks on the transcription of T7 coliphage DNA by Escherichia coli RNA polymerase. Biochem J. 1985 Jul 1;229(1):173–181. doi: 10.1042/bj2290173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques J. P., Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 1991 May;5(5):707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- Klinedinst D. K., Drinkwater N. R. Mutagenesis by apurinic sites in normal and ataxia telangiectasia human lymphoblastoid cells. Mol Carcinog. 1992;6(1):32–42. doi: 10.1002/mc.2940060107. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Schaaper R. M., Loeb L. A. Depurination-induced infidelity of deoxyribonucleic acid synthesis with purified deoxyribonucleic acid replication proteins in vitro. Biochemistry. 1983 May 10;22(10):2378–2384. doi: 10.1021/bi00279a012. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Borden A., Banerjee S. K., LeClerc J. E. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 1990 Apr 25;18(8):2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Preston B. D. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Mamet-Bratley M. D. Transcription in vitro from a DNA template containing apurinic sites. Biochim Biophys Acta. 1974 Mar 27;340(3):237–243. doi: 10.1016/0005-2787(74)90269-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mellon I., Hanawalt P. C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989 Nov 2;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Pieper R. O., Futscher B. W., Erickson L. C. Transcription-terminating lesions induced by bifunctional alkylating agents in vitro. Carcinogenesis. 1989 Jul;10(7):1307–1314. doi: 10.1093/carcin/10.7.1307. [DOI] [PubMed] [Google Scholar]
- Reines D., Wells D., Chamberlin M. J., Kane C. M. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J Mol Biol. 1987 Jul 20;196(2):299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990 Dec 5;265(34):21330–21336. [PubMed] [Google Scholar]
- Selby C. P., Witkin E. M., Sancar A. Escherichia coli mfd mutant deficient in "mutation frequency decline" lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss B., Rabkin S., Sagher D., Moore P. The role of DNA polymerase in base substitution mutagenesis on non-instructional templates. Biochimie. 1982 Aug-Sep;64(8-9):829–838. doi: 10.1016/s0300-9084(82)80138-7. [DOI] [PubMed] [Google Scholar]
- Thrall B. D., Mann D. B., Smerdon M. J., Springer D. L. DNA polymerase, RNA polymerase and exonuclease activities on a DNA sequence modified by benzo[a]pyrene diolepoxide. Carcinogenesis. 1992 Sep;13(9):1529–1534. doi: 10.1093/carcin/13.9.1529. [DOI] [PubMed] [Google Scholar]
- Wada K., Wada Y., Doi H., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):1981–1986. doi: 10.1093/nar/19.suppl.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]