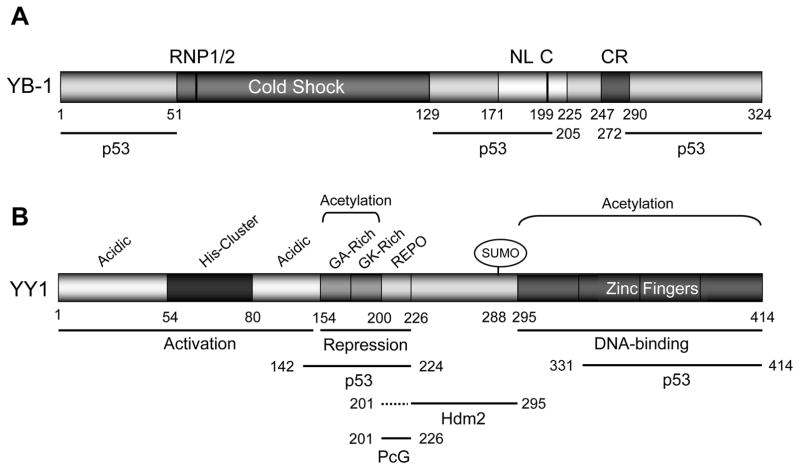
Figure 3. The domain structures of YB-1 and YY1 proteins.
(A) The structure of YB-1. YB-1 has a cold-shock DNA-binding domain with two transactivation domains at both amino- and carboxyl-termini through which it binds to p53. The cold-shock domain has RNP1/2-like motifs consisting of a five-stranded β-barrel structure that creates a surface that may act as a large nucleic acid-binding site. The inactive protein is in the cytoplasm (the cytoplasmic retention [CR] signal is at a.a. 247–290). The nuclear localization signals [NL] have been mapped to a.a. 171–225 by Jürchott et al. (56). This protein needs to be cleaved at a.a. 219–220 [C] before nuclear localization. The p53-binding domains have been mapped to N-terminal, middle, and C-terminal domains (a.a. 1–51, 129–205, and 272–324) (27).
(B) The structure for human YY1. Human YY1 consists of 414 amino acids with amino-terminal transactivation domain containing 11x His cluster with a carboxyl terminal zinc finger domain. The GA, GK-rich, and REPO domains have been shown to be responsible for transcriptional repression. p300 and CBP associated factor (PCAF) mediate the acetylation of residues 171–200 while PCAF also acetylates the C-terminal zinc finger domain of YY1. The REPO domain of YY1 binds and recruits PcG transcriptional repressors. p53 binds to YY1 amino acid residues 142–224, and 331–414; Hdm2 binds to YY1 through amino acid residues 226–295; thus YY1 make a ternary complex with p53 and Mdm2, and inactivating p53-mediated transcription (30).