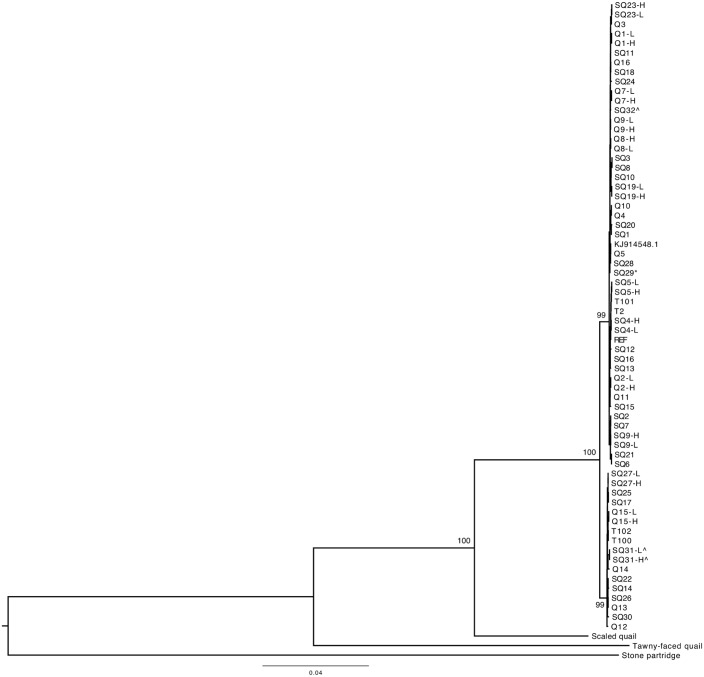
Fig 4. Maximum Likelihood-based Phylogeny Constructed with Expanded Taxon Sampling.
Phylogeny of all bobwhite mitogenomes (n = 66, including 13 heteroplasmic minor allele haplotypes) in conjunction with mitogenomes for the scaled quail (Callipepla squamata), tawny-faced quail (Rhynchortyx cinctus) [37], and stone partridge (Ptilopachus petrosus) [37]. The asterisk (*) denotes a pen-released (n = 1) origin, and ‘^’ denotes bobwhites sampled from active surrogating pastures (i.e., pen release sites; n = 3). Terminal taxa noted with “H” and “L” refers to the high frequency (i.e., major) and low frequency (i.e., minor) heteroplasmic haplotypes, respectively. Individual bobwhites are labeled with laboratory identifiers (Q, SQ, T), with REF indicating the bobwhite reference sequence (GenBank Accession AWGT00000000.1), and KJ914548.1 indicating a bobwhite GenBank Accession [37] included in our analyses. The maximum likelihood phylogeny was constructed with RAxML 7.2.8 [96] using a GTR+Γ model of sequence evolution, with bootstrap support values based on 1,000 pseudoreplicates.