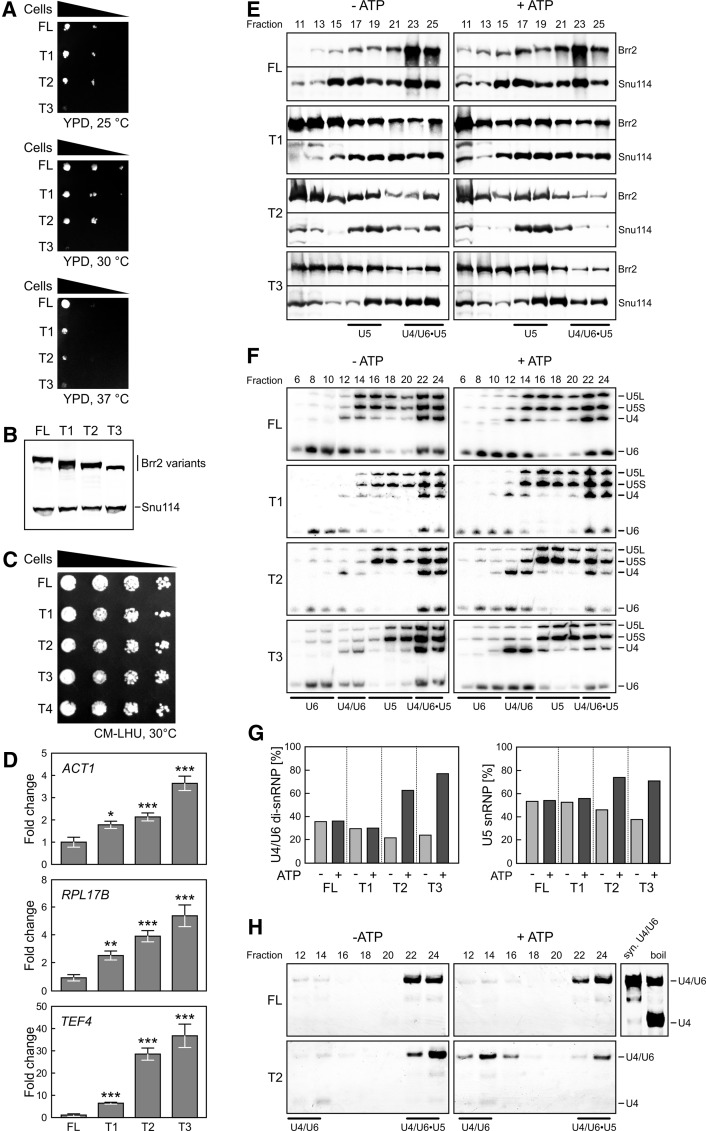
Figure 2.
Physiological effects of Brr2 NTR truncations. (A) Yeast growth assay comparing strains that produce the indicated Brr2 variants (FL, T1, T2, and T3) as the sole type of Brr2 protein. The T4 strain was not viable. (B) Western blot analysis monitoring the types and amounts of Brr2 variants relative to the U5 protein Snu114 produced in extracts of the strains shown in A. (C) Yeast growth assay comparing strains that produce the indicated Brr2 variants (FL, T1, T2, T3, and T4) in a FL Brr2 background. (D) Accumulation of intron-containing transcripts of the indicated genes (ACT1, RPL17B, and TEF4) relative to the amounts of intronless THD1 transcripts in yeast strains producing the indicated Brr2 variants (FL, T1, T2, and T3) as the sole type of Brr2 protein. Values and error bars represent means ± standard error of the mean (SEM) of two technical duplicates of at least four independent experiments. Significance (Student's unpaired t-test) is indicated by asterisks. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (E) Western blots of odd-numbered glycerol gradient fractions of extracts from yeast strains producing the indicated Brr2 variants (FL, T1, T2, and T3). (Left panels) Without pretreatment (− ATP). (Right panels) After preincubation with ATP (+ ATP). Proteins are identified at the right. (F) Northern blots of even-numbered glycerol gradient fractions. snRNPs contained in the fractions are indicated below the gels, and snRNAs are identified at the right. (G) Quantification of the data in F. The left panel shows the amounts of U4 plus U6 snRNAs in U4/U6 di-snRNP fractions relative to U4 plus U6 snRNAs in combined di-snRNP and tri-snRNP fractions (before and after the addition of ATP). The right panel shows the amounts of U5 snRNA in U5 snRNP fractions relative to U5 snRNA in combined U5 snRNP and tri-snRNP fractions (before and after the addition of ATP). (H) Solution hybridization analysis (probing for U4 snRNA) of RNAs extracted from the fractions in F and separated by nondenaturing PAGE. Migration positions of the U4/U6 duplex (syn. U4/U6; obtained by annealing of in vitro transcribed RNAs) and single-stranded U4 snRNA are shown at the right. Boil time was chosen so that part of U4/U6 remained associated.