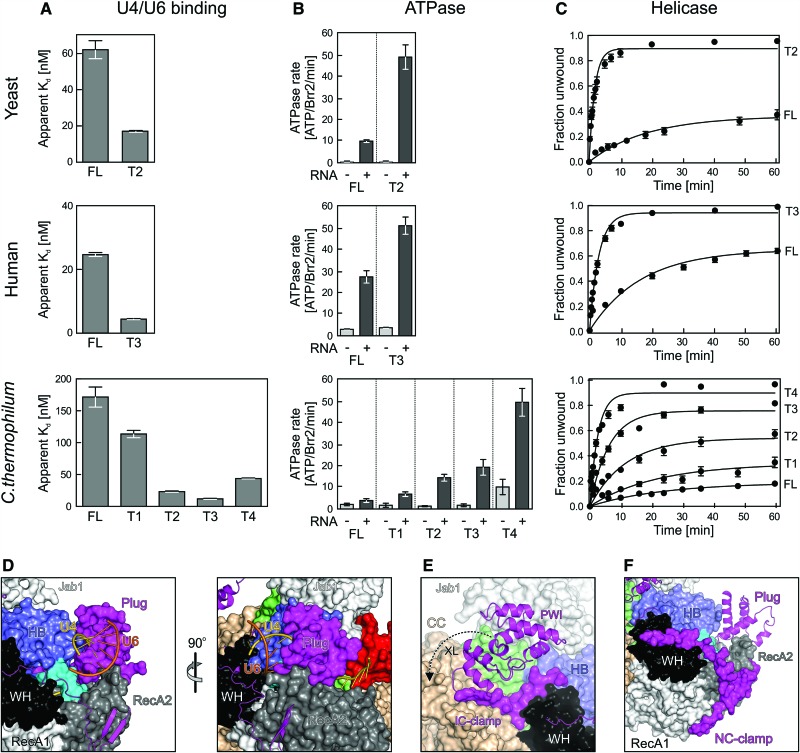
Figure 3.
Effects of NTR truncations on yeast, human, and C. thermophilum Brr2 activities. (A) Apparent Kd values of the indicated Brr2 variants binding to U4/U6 di-snRNAs (organisms are indicated at the left, and protein variants are indicated below the graphs). Values represent means ± SEM of at least two independent experiments. Apparent Kd values were obtained by fitting quantified data from electrophoretic gel mobility shift assays (EMSA) titrations to a single exponential Hill function {fraction bound = A[protein]n/([protein]n + Kd n), where A is the fitted maximum of RNA bound, and n is the Hill coefficient} (Ryder et al. 2008). (B) ATPase activities of the indicated Brr2 variants determined by thin-layer chromatography (organisms are indicated at the left, and protein variants are indicated below the graphs). (−) Intrinsic ATPase activities; (+) U4/U6-stimulated ATPase activities. Values represent means ± SEM of at least three independent experiments. (C) Quantification of U4/U6-unwinding time courses using the indicated Brr2 variants (organisms are indicated at the left, and protein variants are indicated below the graphs). Radioactive bands on gels monitoring the unwinding reactions were quantified by densitometry and fit to a first-order reaction [fraction unwound = A{1 − exp(−ku t)}, where A is the amplitude of the reaction, ku is the apparent first-order rate constant of unwinding, and t is time]. Data points and error bars represent means ± SEM of at least three independent experiments. (D–F) Close-up views of the structure of the FL Brr2–Jab1 complex illustrating the effects of NTR elements removed from the various truncations. (D) Orthogonal views illustrating steric hindrance of U4/U6 di-snRNA binding by the plug. (E) Conformational clamping of N-terminal and C-terminal cassettes by the IC clamp and the PWI domain. (F) Conformational clamping of the N-terminal cassette by the NC clamp. Views in the left panel of D and in E and F are the same as in the top of Figure 1B.