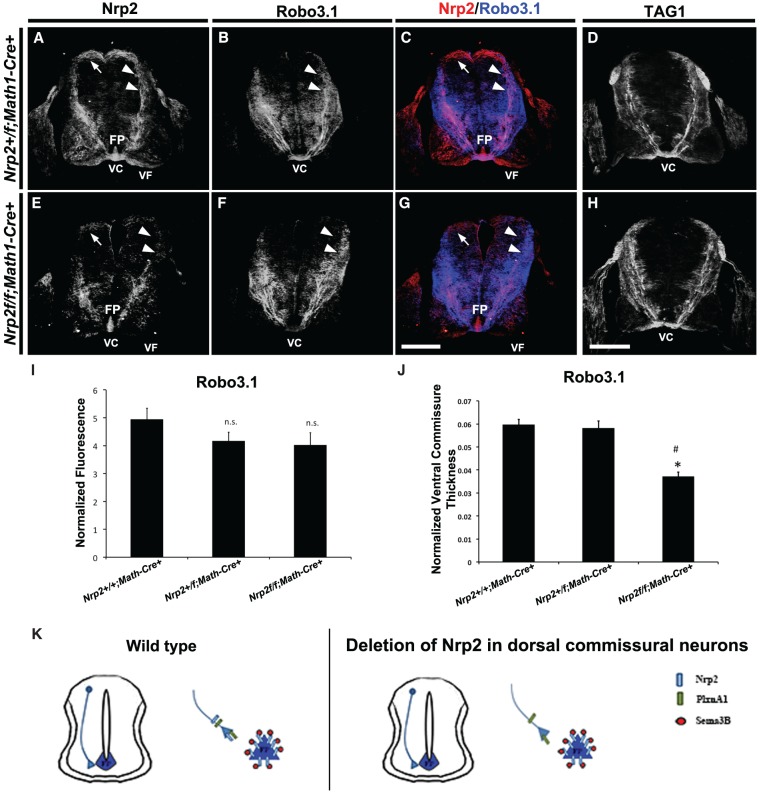
Figure 5.
Specific deletion of Nrp2 from dorsal commissural neurons in Nrp2f/f;Math1-Cre+ spinal cords shows normal precrossing axon guidance. (A–H) Representative confocal micrographs of E11.5 mouse spinal cord sections taken from Nrp2+/f;Math1-Cre+ (A–C) and Nrp2f/f;Math1-Cre+ (E–H) littermate embryos. The same transverse section was processed for immunohistochemistry against Nrp2 (red) and Robo3.1 (blue) in A–C and E–G. Bar in G (for A–C,E–G), 250 µm . (D,H) Representative confocal micrographs of E11.5 mouse spinal cord sections from Nrp2+/f;Math1-Cre+ (D) and Nrp2f/f;Math1-Cre+ (H) immunolabeled using antibodies against TAG1. Bar in H (for D,H), 250 µm. White arrows and white arrowheads point to Nrp2+ cell body location and axons, respectively, in A, C, E, and G. White arrowheads in B and F point to corresponding Robo3.1+ axons relative to Nrp2+ axons. (FP) Floor plate; (VC) ventral commissure; (VF) ventral funiculus. (I) Quantifications of Robo3.1-normalized fluorescence in precrossing axons showed no significant difference between Nrp2+/+;Math1-Cre+, Nrp2+/f;Math1-Cre+ and Nrp2f/f;Math1-Cre+ littermates. Data are means ± SEM from five sections measured per embryo, where n = 5 embryos per genotype. ANOVA, (n.s.) not significant. (J) Quantifications of normalized ventral commissure thickness in Robo3.1-positive precrossing axons in Nrp2+/+;Math1-Cre+ and Nrp2+/f;Math1-Cre+ littermates show no significant difference. A significant decrease in ventral commissure thickness was observed in Nrp2f/f;Math1-Cre+ compared with littermate controls. Data are means ± SEM from five to eight sections measured per embryo, where n = 4 Nrp2+/f;Math1-Cre+ and n = 3 Nrp2f/f;Math1-Cre+. ANOVA, post-hoc Tukey, (*) P < 0.05 compared with Nrp2+/+;Math1-Cre+; (#) P < 0.05 compared with Nrp2+/f;Math1-Cre+. (K) Schematic diagram illustrating wild-type (left) and axonal deletion of Nrp2 (right) genotypes and phenotypes observed in E11.5 spinal cords. Analysis of specific deletion of PlxnA1 in dorsal commissural neurons and PlxnA1 global knockout mutants are shown in Supplemental Figures 6 and 7, respectively.