Abstract
Purpose
Plasma membranes of lens fiber cells have high levels of long-chain saturated fatty acids, cholesterol, and sphingolipids—key components of lipid rafts. Thus, lipid rafts are expected to constitute a significant portion of fiber cell membranes and play important roles in lens biology. The purpose of this study was to characterize the lens lipid raft proteome.
Methods
Quantitative proteomics, both label-free and iTRAQ methods, were used to characterize lens fiber cell lipid raft proteins. Detergent-resistant, lipid raft membrane (DRM) fractions were isolated by sucrose gradient centrifugation. To confirm protein localization to lipid rafts, protein sensitivity to cholesterol removal by methyl-β-cyclodextrin was quantified by iTRAQ analysis.
Results
A total of 506 proteins were identified in raft-like detergent-resistant membranes. Proteins identified support important functions of raft domains in fiber cells, including trafficking, signal transduction, and cytoskeletal organization. In cholesterol-sensitivity studies, 200 proteins were quantified and 71 proteins were strongly affected by cholesterol removal. Lipid raft markers flotillin-1 and flotillin-2 and a significant fraction of AQP0, MP20, and AQP5 were found in the DRM fraction and were highly sensitive to cholesterol removal. Connexins 46 and 50 were more abundant in nonraft fractions, but a small fraction of each was found in the DRM fraction and was strongly affected by cholesterol removal. Quantification of modified AQP0 confirmed that fatty acylation targeted this protein to membrane raft domains.
Conclusions
These data represent the first comprehensive profile of the lipid raft proteome of lens fiber cells and provide information on membrane protein organization in these cells.
Keywords: proteomics, mass spectrometry, lipid raft
Cell membranes are composed of a complex array of lipids and these lipids do not distribute randomly throughout the membrane but, instead, form distinct membrane domains via lipid-lipid and lipid-protein interactions.1 The lipid raft hypothesis was initially proposed in 1997 by Simons and Ikonen2 as a principle of membrane subcompartmentalization. Later in 2006 at the Keystone Symposium on Lipid Rafts and Cell Function, lipid rafts were defined as small, heterogeneous, highly dynamic membrane microdomains (10–200 nm) that are enriched in cholesterol and sphingolipids and that compartmentalize cellular processes.3 As is now increasingly appreciated, lipid rafts may play important roles in various cellular processes, such as signal transduction, membrane trafficking, cell adhesion, cytoskeletal rearrangement, and many other membrane functions.4
The concept of lipid sorting into membrane microdomains was initially introduced to explain the generation of the glycosphingolipid-rich apical membrane in epithelial cells.5 Later, this idea was expanded to include cholesterol-rich raft microdomains.2,6,7 Based on the lipid raft hypothesis,2 sphingolipids associate laterally with one another in the exoplasmic leaflet and any void space is filled with cholesterol. Cholesterol is also present in the cytoplasmic leaflet and fills the void space created by interdigitating fatty acid chains. Cholesterol is known to increase the thickness and stiffness of lipid bilayers and it is generally regarded as a key lipid component of lipid rafts.8
Plasma membranes of lens fiber cells are distinguished from other eukaryotic cell membranes by their unique lipid composition. Lens fiber cell membranes contain high concentrations of long-chain saturated fatty acids and high abundances of sphingomyelin and cholesterol.9–11 This unique lipid composition gives rise to a unique lens fiber cell membrane that is important for lens transparency.11–13 The abundance of sphingomyelin and cholesterol contributes to lens membrane rigidity and structural order. Considering this unusually high concentration of cholesterol and sphingolipid, it is expected that lipid raft domains are highly abundant and play important roles in lens fiber cell membrane. Evidence for immiscible cholesterol-rich and -poor domains in normal lens fiber cell membrane was reported,14 and cholesterol-rich domains were more pronounced and better defined in cataractous lens.15 Previously, lipid raft domains were isolated and lipids13 and caveolin-116 content was examined. Sorting of AQP0 and connexins to raft and nonraft domains was also reported.17 However, the structure and function of lipid rafts in lens fiber cells are largely unknown.
Lipid rafts have been isolated from a variety of tissues relying on their insolubility in nonionic detergents at low temperature. Although detergent insolubility in itself does not accurately reflect preexisting raft domains, studying detergent-resistant membranes (DRMs) has produced results consistent with other methods, such as direct imaging.18,19 Thus, detergent resistance remains an interesting and useful tool for assigning potential membrane raft association. In this report, we isolated DRM from bovine lens fiber cells and performed quantitative proteomic studies of DRM and detergent-soluble membrane (DSM) fractions. Our results demonstrated that a significant percentage of AQP0, MP20, AQP5, and a small fraction of connexin 50 and connexin 46 reside in lipid raft domains.
Materials and Methods
Bovine lenses (1 year or older) were obtained from PelFreez Biologicals (Rogers, AK, USA). Sequence-grade modified trypsin was obtained from Promega (Madison, WI, USA). Brij 98 was purchased from Acros Organics (Morris Plains, NJ, USA). Methy-β-cyclodextrin and other chemicals were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). All HPLC grade solvents were purchased from Fisher (Fair Lawn, NJ, USA).
Preparation of Detergent-Resistant Membrane
Frozen bovine lenses were decapsulated and dissected into cortex (the superficial soft tissue) and nucleus (the remaining hard gummy tissue) before homogenization. Tissue was homogenized in homogenizing buffer (200 mM sucrose, 150 mM NaCl, 50 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF] in 10 mM Hepes) and centrifuged at 100,000g for 20 minutes and the supernatant was discarded. The pellets were resuspended in homogenizing buffer, divided into two samples, and centrifuged at 100,000g for 20 minutes; 1 mL homogenizing buffer, with or without 15 mM or 35 mM methyl-β-cyclodextrin (MβCD), was added to the pellets, and the samples were vortexed and incubated at 37°C for 30 minutes. The samples were then further centrifuged and the pellets were collected; 500 μL 1% Brij 98 and 15 mM octylglucoside (OG) in homogenizing buffer was added to each pellet and incubated on ice for 30 minutes. Each sample was then mixed with 500 μL 83.2% sucrose in homogenizing buffer containing 1% Brij 98 to make final sucrose concentration of 45% and loaded at the bottom of the centrifuge tube. Thirty-five percent and 5% sucrose prepared in homogenizing buffer were layered on the top of the sample sequentially and centrifuged at 160,000g using a SW55Ti rotor in a Beckman L90K ultracentrifuge (Fullerton, CA, USA) for 18 hours at 4°C. Ten fractions (0.5 mL/fraction) were collected across the sucrose gradient and fractions 3 to 10 were used for further analysis.
Preparation of Samples for MS Analysis
Proteins in 100 μL each fraction were precipitated by chloroform/methanol and the resulting protein pellets were solubilized in 1% SDS for protein assay. The total protein concentration was measured by a bicinchoninic acid assay (BCA assay; Thermo Scientific, Rockford, IL, USA). For proteomic analysis, 200 μL from each fraction was reduced with DTT (final concentration of 10 mM) at 56°C for 1 hour and alkylated with iodoacetamide (final concentration 50 mM) at room temperature for 45 minutes. The samples were concentrated in a Speedvac to 100 μL and proteins in each sample were then precipitated by chloroform/methanol. The resulting proteins were suspended in 10% acetonitrile in 50 mM Tris buffer, pH 8.0. Trypsin was added (enzyme/substrate = 1/100; Promega), and the sample was digested at 37°C for 18 hours. After digestion, the sample was dried by a speedvac (Thermo Fisher, Milford, MA, USA) and reconstituted in 0.1% formic acid. The supernatant was collected by centrifuging at 20,000g for 5 minutes and the remaining pellets were extracted by 50 μL 99.9% acetonitrile (ACN), 0.1% formic acid. The ACN extract was dried by a speedvac and reconstituted in 5% ACN (0.1% formic acid). Equal volumes of ACN extract and 0.1% formic acid extract were mixed before liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
For isotope-coded tags for relative and absolute quantitation (iTRAQ) analysis, fractions 3 and 4 were diluted 15 times by water and centrifuged at 200,000g for 30 minutes. The pellets were washed twice with water and reconstituted in 500 mM triethylammonium bicarbonate (TEAB; pH 8.0) containing 10% ACN. Proteins were reduced with 50 mM TCEP (Tris-[2-carboxyethyl]phosphine) at 60°C for 1 hour, alkylated with 200 mM MMTS (methyl methanethiosulfonate) at room temperature for 10 minutes, and digested with sequencing-grade trypsin overnight. Peptides were then labeled with iTRAQ reagents according to the manufacturer's instructions (AB Sciex, Foster City, CA, USA). Labeling reagents were reconstituted in ethanol such that each protein sample was labeled at a final concentration of 90% ethanol and labeling was performed for 2 hours. The 35 mM MβCD-treated sample was labeled with the 114 iTRAQ reagent and the untreated sample was labeled with 116 iTRAQ reagent. The iTRAQ-labeled MβCD-treated sample and one-fifth of iTRAQ-labeled untreated sample were mixed, acidified with trifluoroacetic acid, and were subsequently desalted by a modified Stage-tip method20 before LC-MS/MS analysis.
Liquid Chromatography–Electrospray Ionization/MS/MS Analysis
Tryptic peptides were either directly separated on a one-dimensional fused silica capillary column (20 cm × 100 μm) packed with Jupiter resin (3-μm mean particle size, 300 Å pore size; Phenomenex, Torrance, CA, USA) or analyzed by MudPIT.21 One-dimensional liquid chromatography was used with the following gradient at a flow rate of 0.5 μL/min: 0 to 2 minutes: 2% ACN (0.1% formic acid), 2 to 70 minutes: 2% to 35% ACN (0.1% formic acid), 70 to 90 minutes: 35% to 90% ACN (0.1% formic acid) balanced with 0.1% formic acid. The eluate was directly infused into a Velos Pro linear ion trap mass spectrometer (ThermoFisher, San Jose, CA, USA) equipped with a nanoelectrospray source. For MudPIT analysis, peptides were loaded onto a custom packed biphasic C18/SCX trap column (4 cm × 150 μm, Jupiter C18, 5 μm, 300 Å media followed by 4 cm × 150 μm, Luna SCX, 5 μm, 100 Å media; Phenomenex). The trap column was coupled to a capillary analytical column (20 cm × 100 μm, Jupiter C18, 3 μm, 300 Å media). MudPIT analysis was done with a 13-step salt pulse gradient (0 mM, 25 mM, 50 mM, 75 mM, 100 mM, 150 mM, 200 mM, 250 mM, 300 mM, 500 mM, 750 mM, 1 M, and 2 M ammonium acetate). Following each salt pulse, peptides were eluted from the analytical column with a 90-minute reverse-phase solvent gradient (2%–45% ACN, 0.1% formic acid; 2%–95% ACN, 0.1% formic acid for last two salt pulses). The eluate was directly electrosprayed into a Velos Pro mass spectrometer (ThermoFisher). The instrument was operated in a data-dependent mode with one precursor scan event to identify the top 15 most abundant ions in each MS scan, which were then selected for fragmentation. Dynamic exclusion (repeat count 1, exclusion list size 300, and exclusion duration 15 seconds) was enabled to allow detection of less abundant ions. For quantification of fatty acylated AQP0, the samples were run on a one-dimensional column as above with modified gradient (0–50 minutes: 2%–30% ACN, 50–65 minutes: 30%–95% ACN, 65–80 minutes: 95% ACN balanced with 0.1% formic acid) and the mass spectrometer was set to acquire one MS1 scan followed by targeted MS/MS scans for fatty acylated AQP0 peptides and unmodified AQP0 peptides. The iTRAQ-labeled samples were analyzed using MudPIT analysis as described above with eight salt pulse steps (0, 50 mM, 100 mM, 200 mM, 300 mM, 500 mM, 1 M, and 2 M ammonium acetate). Peptides were introduced via nano-electrospray into a Q Exactive mass spectrometer (Thermo Scientific, San Jose, CA, USA). The Q Exactive was operated in data-dependent mode acquiring higher-energy collisional dissociation (HCD) MS/MS scans (R = 17,500) after each MS1 scan (R = 70,000) on the 18 most abundant ions using an MS1 ion target of 1 × 106 ions and an MS2 target of 1 × 105 ions. The maximum ion time for MS/MS scans was set to 100 ms, the HCD-normalized collision energy was set to 26, dynamic exclusion was set to 30 seconds, and peptide match and isotope exclusion were enabled.
Data Analysis
Tandem mass spectra were analyzed using a suite of custom-developed bioinformatics tools. All MS/MS spectra were converted to mzML files by Scansifter, a tool under development at Vanderbilt University Medical Center, and searched on a 2500-node Linux cluster supercomputer using a custom version of the TagRecon algorithm.22 Trypsin specificity was used with a maximum two missed cleavage sites. The data were searched against a Uniprot bovine database (October 18, 2013) with a static modification of carbamidomethylation of cysteine residues and variable modifications of oxidation of methionine and deamination of N-terminal glutamine residues. The search results were filtered by IDPicker23 by controlling protein false-discovery rate (FDR) to less than 1%. For quantitative peak area measurements, selected ion chromatograms were generated and peaks areas were calculated using the Genesis peak algorithm within the Xcalibur software 2.2 SP 1.48 (ThermoFisher). For quantification of the relative abundance of fatty acylated AQP0, the resulting raw files were imported into Skyline24 for peak-picking and quantitation. At least seven product ions were used for each peptide and the total peak area from all product ions selected was normalized to the total peak area from all product ions of the AQP0 peptide 188 to 196. For iTRAQ data analysis, mass spectra were processed using the Spectrum Mill software package (version B.04.00; Agilent Technologies, Santa Clara, CA, USA). The MS/MS spectra acquired on the same precursor m/z (±0.01 m/z) within ±1 second in retention time were merged. The MS/MS spectra of poor quality that failed the quality filter by not having a sequence tag length greater than 1 were excluded from searching. A minimum matched peak intensity requirement was set to 50%. For peptide identification, MS/MS spectra were searched against a Uniprot bovine database (Oct 18, 2013). Additional search parameters included trypsin enzyme specificity with a maximum of three missed cleavages, ±20 ppm precursor mass tolerance, ±20 ppm (HCD) product mass tolerance, and fixed modifications including MMTS alkylation of cysteines and iTRAQ labeling of lysines and peptide N-termini. Oxidation of methionine was allowed as a variable modification. Autovalidation was performed such that peptide assignments to mass spectra were designated as valid following an automated procedure during which score thresholds were optimized separately for each precursor charge state and the maximum target-decoy–based FDR was set to 1.0%. To obtain iTRAQ protein ratios, which represent control/MβCD-treated ratios, the median was calculated for all peptides assigned to each protein. Because only one-fifth of the control sample was used, the final iTRAQ ratio was obtained by multiplying by a factor of 5.
Results
Isolation of Lipid Raft-Like DRMs
The most commonly used method for isolation of raft-like DRMs uses the nonionic detergent Triton X-100; however, sucrose gradient centrifugation of the lens Triton X-100–resistant fraction yields only trace amounts of DRM from lens fiber cell membrane.16 Consistently, we found that there was no visible band at the 5% to 35% sucrose interface if the fiber cell membrane was solubilized with 1% Triton X-100. Therefore, octyl-β-glucopyranoside (OG) was included in the preparation as previously reported.16 In addition, Mg2+ and K+ ions were included in the detergent solubilization buffer as described previously by Chen et al.25 to mimic the intracellular environment and stabilize the inner leaflet of the membranes during detergent solubilization.
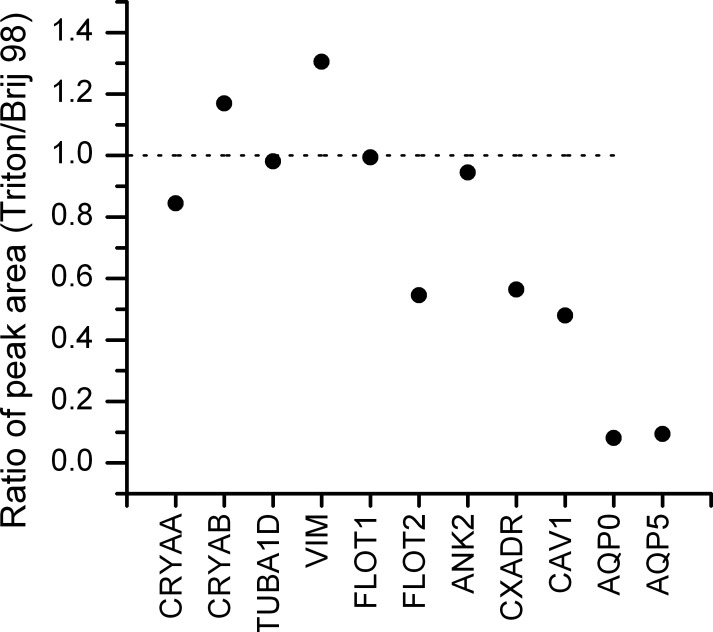
Even though Triton X-100 was the preferred detergent for membrane raft isolation, Triton X-100 was also proposed to potentially induce the artificial formation of detergent-resistant structures.26 A milder detergent, the polyoxyethylene ether Brij 98, has been used for solubilizing nonraft domains because its mono-unsaturated ether moiety was predicted to preferentially solubilize loosely packed fluid phase lipids.27,28 Advantages of Brij 98 include isolation of lipid rafts at 37°C and reduced aggregation of detergent-insoluble membranes following detergent extraction. Thus, we tested both Triton X-100 and Brij 98 in combination with OG for isolation of DRM. The fraction at the 5% to 35% sucrose interface was collected from both preparations, and proteins in this fraction were analyzed by MS. Figure 1 shows the relative abundance of several abundant lens proteins and raft marker proteins in the Triton X-100–treated sample compared with the Brij 98–treated sample. The Brij 98–treated sample yielded higher amounts of raft marker proteins (FLOT1, FLOT2, ANK2, CXADR, CAV1) in the high-buoyancy fraction compared with the Triton X-100–treated sample, but did not significantly increase the presence of soluble putative nonraft proteins, such as crystallins (CRYAA, CRYAB), and cytoskeletal proteins, such as vimentin and tubulin. In addition, the Brij 98 treatment dramatically increased the amount of AQP0 and AQP5 in the low-density fraction. Increasing evidence supports the lipid raft localization of AQP5,29–31 whereas the localization of AQP0 in lipid rafts has been reported previously.16,17 These results suggest that Brij 98 provides better yields of lipid raft marker proteins in the low-density fractions; therefore, further studies were performed only on DRM isolated after Brij 98 solubilization.
Figure 1.
The relative protein abundance in DRM fractions prepared using 1% Triton X-100 or 1% Brij 98. The DRMs were prepared using either 1% Triton X-100 plus 15 mM OG or 1% Brij 98 plus 15 mM OG as described in the Materials and Methods section. Proteins at the 5% to 35% sucrose interface were digested by trypsin and analyzed by LC-MS/MS. The abundance of each protein was represented as the peak area of one peptide from this protein. The relative abundance of a certain protein in two different detergent-treated samples was plotted as a ratio of the peak areas.
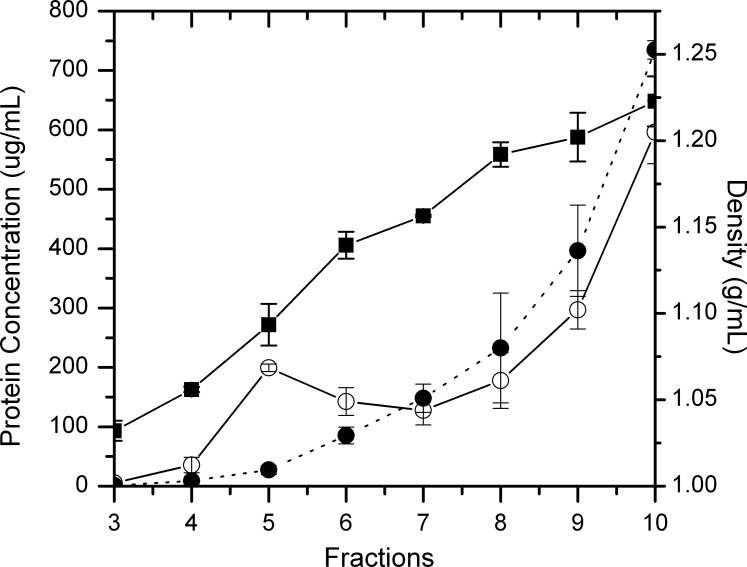
In the course of method optimization, we noticed that the detergent-protein ratio affects the amount of proteins in the high buoyancy fractions (data not shown). During detergent solubilization, the total protein concentration was controlled to be 3 mg/mL. Under this condition, the total protein concentration of different fractions collected across the sucrose density gradient is shown in Figure 2. Fractions 4 and 5 correspond to the 5% to 35% sucrose interface. Only a small fraction of the total proteins was present in the high-buoyancy fractions (fractions 3–5); however, the detergent-soluble fractions were dominated by highly abundant soluble crystallins. As expected, removing cholesterol by MβCD significantly reduced the yield of raft-like DRM and shifted proteins from the DRM to the DSM.
Figure 2.
The total protein concentration in different fractions collected from raft-like DRM preparations using 1% Brij 98 plus 15 mM OG. Proteins in 100 μL each fraction were precipitated by chloroform/methanol. The resulting pellets were solubilized in 1% SDS and protein concentration was measured by BCA assay. The average protein concentration of each fraction from two experiments was plotted. Open circles: protein concentration; filled circles: protein concentration after MβCD treatment; filled squares: density.
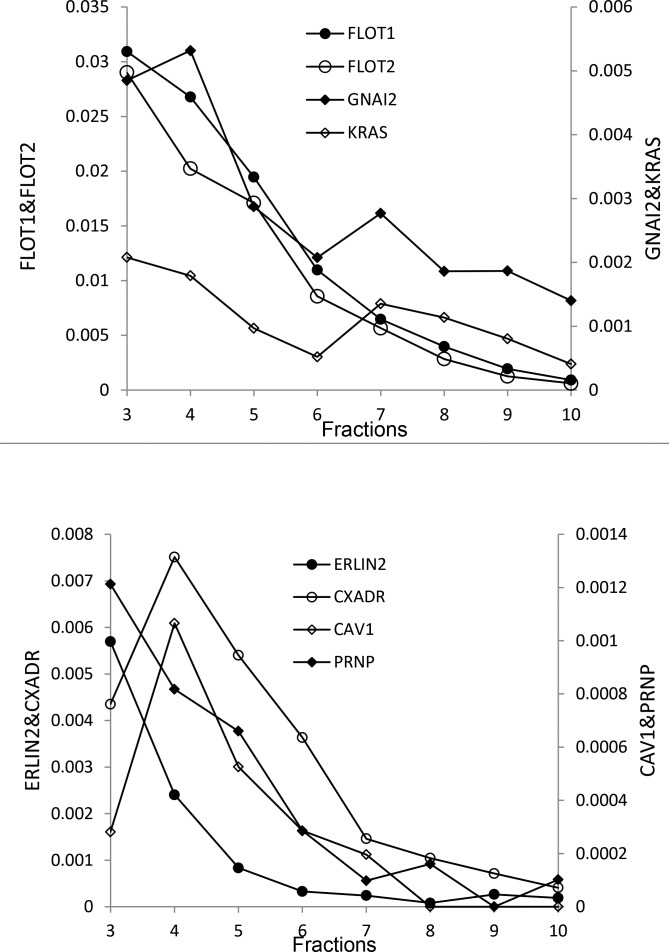
To evaluate whether our preparation yielded bona fide raft-like DRM, the distribution of several lipid raft markers across different sucrose density gradient was studied. The relative abundance of a protein in each fraction was represented by its normalized spectral count in the LC-MS/MS data; a rough estimate of protein abundance.32 This experiment was repeated three times. Consistently, many lipid raft markers were repeatedly detected in the DRM faction, such as flotillin 1 and 2, erlin 1 and 2, G proteins, caveolin 1 and 2, HRAS, and KRAS. Using this preparation, most of these proteins are highly enriched in lipid raft-like low-density DRM, and are much less abundant or not detected in the DSM fractions. The normalized spectral counts for some raft markers in different fractions showed a similar trend as shown in Figure 3. Some of these proteins, such as KRAS, and GNAI2, have bimodal distributions, suggesting their presence in both DRM and DSM membranes.
Figure 3.
Distribution of lipid raft markers across sucrose density gradient. Proteins in each fraction were digested by trypsin and analyzed by LC-MS/MS. The data were searched against a Uniprot bovine database. The spectral count from each protein was normalized to the total spectral count and the relative abundance of each protein was represented using the normalized spectral count. The y-axis shows the normalized spectral counts for some raft marker proteins.
Identification of Lens Lipid Raft-Like DRM Proteins by LC-MS/MS
To analyze proteins that are present in the high-buoyancy DRM fractions, fractions 3 and 4 were pooled, digested by trypsin, and analyzed by MudPIT. Fraction 5 typically had higher protein concentrations than fraction 4 and contained significant amounts of raft marker proteins; however, the normalized spectral counts for most raft markers were lower in fraction 5 than fraction 4, indicating contamination with nonraft proteins. Therefore, only fractions 3 and 4 were used for identifying proteins in rafts to reduce contamination by nonraft proteins. This experiment was repeated twice. In total, there were 598 proteins detected in the pooled high-buoyancy lipid raft fraction (Supplementary Table S1) and 506 proteins (84%) detected in both analyses. Functional and localization annotations were assigned for 506 proteins that were identified in both analyses using the information in the Uniprot database (http://www.uniprot.org, in the public domain) as well as after analysis by the DAVID Bioinformatic Resources.33 A search for membrane raft proteins in the Uniprot database yielded 251 unique proteins and 40 of these proteins were detected in the DRM fraction. These proteins are listed in the Table 1. Among the detected 506 proteins, 79 proteins are listed as lipidated proteins and 140 proteins (27.7%) were listed as glycosylated proteins in the Uniprot protein database (shown in the Supplementary Table S1). Recently, a mammalian raft proteome database was established based on multiple proteomic studies in a variety of tissue or cells.34 In this database, a high-confidence lipid raft protein list was generated to include only proteins that are sensitive to cholesterol depletion or proteins that are detected in raft domains by more than one biochemical method.34 A comparison of the proteins detected in this study with the proteins in the raft proteome database revealed that 70.9% (359 proteins) of the proteins detected in the DRM of lens fiber cells are present in the high-confidence list. Another 14.8% (75 proteins) can be found in the raft proteome full list including some well-studied raft proteins, such as AQP5, ANK2, KRAS, and STX2. Seventy-two proteins from our lens study are not present in the lipid raft database, including 15 proteins that are uniquely expressed in the lens.
Table 1.
Lipid Raft Proteins Detected in DRM Fractions of Lens Fiber Cells

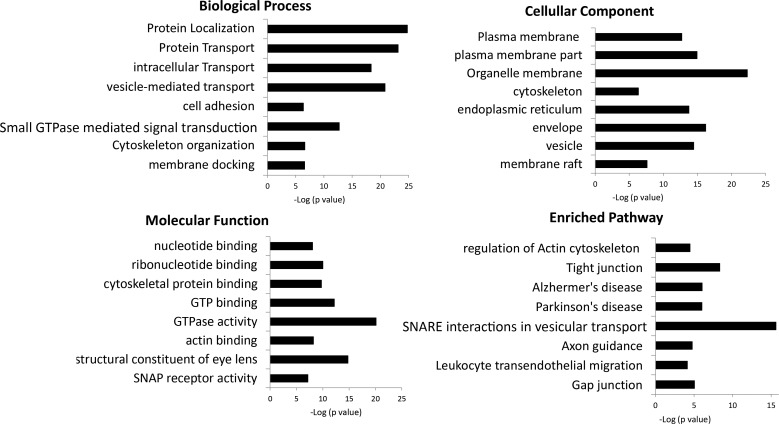
Pathway analysis was also carried out using DAVID Bioinformatic Resources.33 The top enriched GO terms and pathways are listed in Figure 4. Combing both cell membrane and organelle membrane lists, most proteins (75%) detected in our study localize to membranes. Similar to previous reports, mitochondrial and endoplasmic reticulum (ER) proteins were also detected.35–37 Results from enriched biological process and pathway analysis provides evidence that supports important raft-related functions, such as membrane transport and trafficking,38,39 signal transduction,40,41 cell junctions,42,43 and cytoskeleton organization.41,44
Figure 4.
Enriched GO categories and pathways. Enriched GO categories and pathways in raft-like DRM fractions were analyzed using David Bioinformatics Resources; P < 0.0001 was considered enriched GO categories. Selected enriched categories were plotted with categories that have more proteins on the top.
Cholesterol Dependence
Lens fiber cell plasma membranes are unique due to their extremely high relative content of cholesterol.10,45 Cholesterol represents up to 40% of the total lipid10 and the cholesterol-to-phospholipid molar ratio ranges from 1 to 4 in the lens plasma membrane.16,45 Both 5 mM and 10 mM MβCD treatment, commonly used to remove cholesterol, removed only a small fraction of cholesterol from lens fiber cell membranes as evidenced by the presence of the high-buoyancy band at the 5% to 35% sucrose interface (data not shown). Therefore, a series of concentrations of MβCD was tested for cholesterol removal. The results indicate that cholesterol can be depleted with 35 mM MβCD treatment under our experiment conditions. Considering that MβCD treatment could remove cholesterol in nonraft domains46 and 15 mM MβCD treatment can remove the high-buoyancy band at the 5% to 35% sucrose interface, 15 mM MβCD was used for most of our studies; 35 mM MβCD was used for iTRAQ experiments to ensure removal of any residual lipid raft cholesterol.
To quantify the effect of cholesterol removal on the protein content of the DRM fraction and, thereby, validate the lens lipid raft proteome, a quantitative proteomics approach was taken. Similar to a previous report from the M. Mann group,47 the iTRAQ method was used. Samples were extracted with 35 mM MβCD or homogenizing buffer (control) before detergent solubilization. Pooled sucrose gradient fractions 3 and 4 were digested and analyzed. Liquid chromatography MS/MS analysis was used to produce sequence-specific ions for protein identification and iTRAQ reporter ion signals for quantification; 200 proteins were quantified and the detailed list can be found in Supplementary Table S2. Among the 200 proteins quantified, 170 were detected in the label-free MudPIT analysis. Plotting the control/MβCD-treated ratio did not show distinct discontinuities in iTRAQ ratios as previously reported47 (Supplementary Fig. S1); however, the top 71 proteins had control/MβCD-treated ratios greater than 25 and included important lipid raft markers such as flotillins, KRAS, STX2, ezrin, integrin, and major prion protein. Table 2 lists the top 71 proteins as well as their predicted cellular localization and modifications. Most of these proteins localize to plasma membrane and some of them are cytoskeletal proteins that are modified by glycosylation or lipidation. Many proteins in this list are involved in cell junctions. The sedimentation of several major lens proteins present in the high-buoyancy, low-density DRM fractions, including AQP0 and connexins, was very sensitive to cholesterol removal. Furthermore, although peptides from lens membrane intrinsic protein MP20 (LIM2) were not detected by MudPIT analysis, a complementary one-dimensional LC-MS/MS experiment showed MP20 had a higher-control/MβCD-treated ratio than AQP0, indicating a higher sensitivity to cholesterol removal (data not shown).
Table 2.
Proteins With iTRAQ Ratios Greater Than 25, Their Cellular Localization, and Modifications

Table 3 lists the proteins that have a control/MβCD-treated ratio less than 15. The proteins in this list mainly localize to cytoplasm and organelle membranes, such as nucleus, mitochondrion, ER, and Golgi membranes. Two widely used nonraft marker proteins, calnexin and transferrin receptor protein 2, appear in Table 3. Proteins in Table 3 are weakly affected by cholesterol removal and, therefore, are most likely nonraft proteins that cosedimented with raft proteins. Proteins that have control/MβCD-treated ratios between 15 and 25 include some raft marker proteins such as Ankyrin-2, several G proteins, and Rab family proteins (RAB9A, RALB, RAP2B). Several proteins involved in SNARE interaction in vesicular transport are also in this group, such as SNAP23, STX7, VAMP2, and VAMP5. This group also includes some major lens-soluble proteins, such as alpha B crystallin and several beta crystallins. We interpret the presence of these soluble crystallins in lipid raft fractions and with control/MβCD-treated ratios that are somewhat sensitive to cholesterol removal as most likely due to association with lipid raft proteins.
Table 3.
Proteins With iTRAQ Ratios Less Than 15, Their Cellular Localization, and Modifications

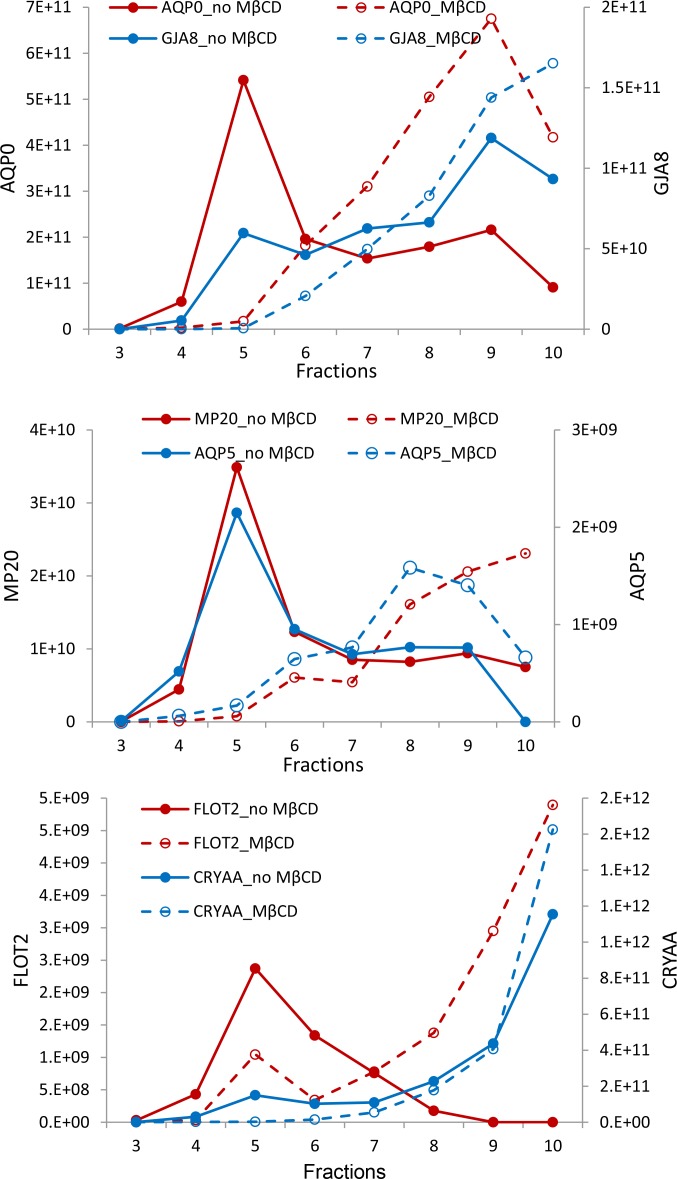
The iTRAQ experiment provided information about how cholesterol removal affects the amount of proteins in raft-like DRM fractions. We also used a label-free, peak area approach to detect the shifts in protein sedimentation throughout the sucrose gradient on cholesterol removal. The abundance of each protein was represented as the peak area of one unique peptide from this protein in each sucrose gradient fraction. Figure 5 shows the abundance of some important lens proteins. This experiment was repeated a second time and the same trend was detected (data not shown). The lipid raft markers flotillin 1 and 2 are primarily detected in the DRM fractions without MβCD treatment; however, they shift to the higher-density DSM fractions on MβCD treatment. Even though the signal for αA crystallin in DRM fraction is strong, the amount of αA crystallin in the DRM fraction was only a very small fraction of total αA crystallin in the sample. A significant percentage of total AQP0, MP20, and AQP5 is present in the low-density DRM fractions, and, after MβCD treatment, protein abundances in the DRM fractions decreased dramatically. Note that some fraction of AQP0, AQP5, and MP20 are detected in the higher-density DSM fractions without MβCD treatment. A lower percentage of connexin 50 (GJA8) is present in the DRM fractions compared with AQP0; however, this portion of connexin 50 shifts to the DSM fraction on MβCD treatment. Connexin 46 followed the same trend as connexin 50 (data not shown).
Figure 5.
Distribution of some major lens proteins across the sucrose density gradient and effects of MβCD treatment. Proteins in each fraction were digested by trypsin and analyzed by LC-MS/MS as described in the Materials and Methods section. Protein abundance was represented as the peak area of one peptide. Total peak area corresponding to proteins from 100 μL each fraction was calculated and plotted. The y-axis shows the peak areas of corresponding peptides.
Fatty Acylation of AQP0
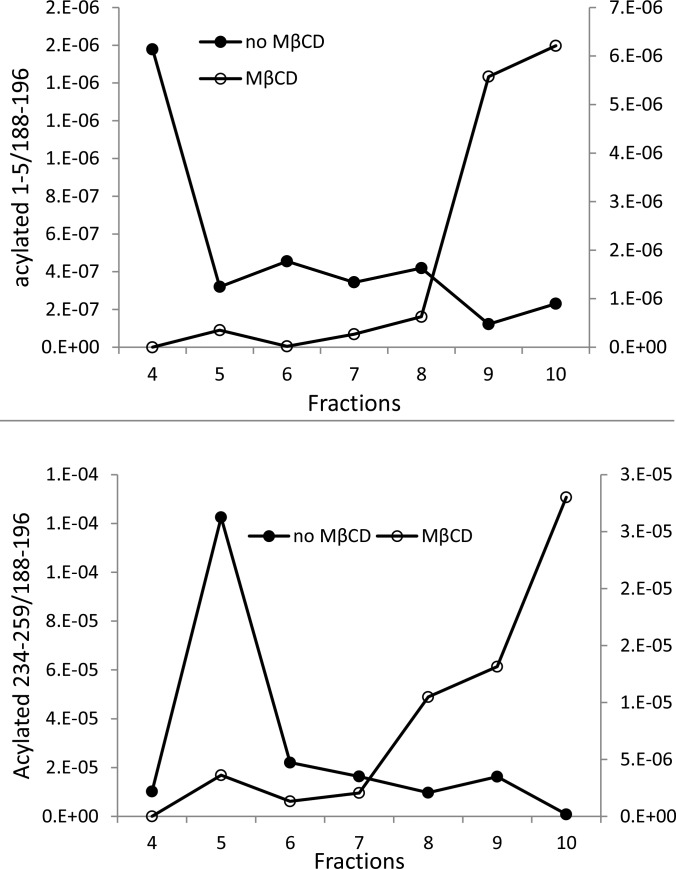
Previously we reported that AQP0 undergoes palmitoylation and oleoylation on Met1 and K238 residues through an amide bond and that modified AQP0 cannot be extracted by Triton X-100, suggesting its possible localization with lipid raft domains.48 In this study, we quantified the level of oleic acid–modified AQP0 relative to total AQP0 signal across the sucrose gradient (Fig. 6). The result shown in Figure 6 is an average of three analyses of three sets of samples. Oleic acid–modified (acylated) AQP0 on Met1 and K238 was highly enriched in fraction 4 and fraction 5, respectively. After MβCD treatment, fatty acylated AQP0 also moved to the DSM fractions. This result further confirmed that fatty acylation of AQP0 acts as one of the signals to target AQP0 to lipid raft domains.
Figure 6.
Quantification of oleic acid–modified AQP0 in different fractions. Chloroform/methanol-precipitated proteins from each fraction were washed with 4 M urea followed by trypsin digestion and analysis by LC-MS/MS as described in the Materials and Methods section. The raw data were imported into Skyline24 for peak-picking and quantification. The total peak area from at least seven product ions was used for each peptide. Signal from the oleic acid–modified AQP0 peptides (peptide 1–5 or 234–259) were normalized by the signal for AQP0 peptide 188 to 196.
Discussion
Methodologic advances continue to provide compelling evidence to support the important functions of lipid rafts; however, lipid raft characterization remains challenging because individual lipid domains cannot be isolated in a pure form. The DRM fraction has been widely used for studying proteins targeting to lipid raft-like domains. Given the high concentration of cholesterol in lens tissue, the detected cholesterol-rich domains in lens fiber cell membranes, and the effects of cholesterol on membrane protein function, it is important to understand the details of membrane protein environments in the lens. In this work, we optimized methods for studying raft-like DRMs from bovine lens fiber cells and our isolation method revealed proteins whose solubility is highly sensitive to MβCD treatment. In this study, 71 proteins were quantified by iTRAQ to be highly sensitive to cholesterol. These proteins included AQP0, AQP5, MP20, and connexins. Previously, AQP0 was reported to localize to both raft and nonraft domains,17 and our results are consistent with this finding. A significant portion, approximately 40%, of the total AQP0 sedimented to DRM fractions. A previous study indicated AQP0 homo-oligomerization is a key factor that targets AQP0 to lipid raft domains.17 Later, AQP0 was found to be modified by fatty acids, such as oleic acid and palmitic acid, and that fatty acylation acted as a potential raft-targeting modification.48 In this study, we confirmed that fatty acylated AQP0 was enriched in the raft-like DRM domains; however, a significant amount of AQP0 in the DRM was not modified by fatty acids, suggesting that other mechanisms exist to target AQP0 to membrane rafts, possibly AQP0 oligomerization. In addition, the presence of a cholesterol-binding or sphingolipid-binding sequence of transmembrane proteins can increase their concentration in lipid raft.49 The AQP0 has four CRAC/CARC sequences that have the potential to bind with cholesterol.50 Further study is needed to confirm whether AQP0 directly binds with cholesterol. A previous report showed that AQP0 water permeability was affected by membrane lipid composition and that increased cholesterol content decreased AQP0 water permeability.51 This result also suggested a direct AQP0-cholesterol interaction.
Among the major lens proteins, MP20 was found to be highly sensitive to cholesterol removal; MP20 is a member of the PMP22/EMP/MP20/Claudin family, a diverse group of proteins with roles in intercellular adhesion and protein trafficking.52,53 The PMP22 is a known constituent of lipid rafts in peripheral nerve myelin and may play a role in the linkage of the actin cytoskeleton with the plasma membrane, likely through regulating the cholesterol content of lipid rafts.54,55 Localization of lens MP20 in lipid rafts has not been reported. Previously, MP20 was found to form large oligomers in the presence of native lens lipids.56 Oligomerization, as reported for AQP0, could target MP20 to lipid raft domains. In addition, almost all tetraspanins are modified by the posttranslational addition of palmitate to membrane-proximal cysteine residues.57 Palmitoylation of MP20 has not been reported, but CSS-Palm software58 predicts palmitoylation of cysteine 11 in MP20. In addition, MP20 also has a CRAC/CARC sequence that could directly interact with cholesterol.50 The MP20 is found in the cytoplasm of young fiber cells and traffics to the plasma membrane concomitantly with the loss of intracellular organelles in maturing lens fiber cells.59 Because lipid raft domains play important roles in protein trafficking and cell adhesion, understanding how MP20 targets to lipid raft domain will help to elucidate the function of this protein in the lens.
Our results suggest that significantly lower amounts of connexins reside in DRM domains compared with AQP0, a result consistent with a previous report that connexins are located primarily in nonraft domains.17 The small fraction of total connexins present in the high-buoyancy DRM was strongly cholesterol sensitive. The observation of two forms of connexins in lens fiber cells, DRM and DSM residing, is supported by a report of cholesterol-rich gap junction and cholesterol-free connexons.60 Connexin family members have been shown to interact with caveolin-1 and to target to lipid raft domains to regulate gap junctional intercellular communication43,61,62; however, previous results showed inconsistency related to the targeting of Cx46 and Cx50 to lipid raft domains.17,61,62 Based on our experiments, the overall distribution of Cx46 and Cx50 across the sucrose gradient was similar, with most of the protein present in the nonraft DSM fraction; however, the small fraction present in the DRM fraction was sensitive to MβCD treatment.
The αA-crystallin was among the proteins that are sensitive to MβCD treatment, whereas most β- and γ-crystallins have much lower MβCD sensitivity. Previously, α-crystallin was the only crystallin reported to noncovalently bind with bovine lens membranes,11,63 and this membrane association property of α-crystallin was attributed to either direct lipid binding64 or association with AQP0.63,65
Compelling evidence supports the central role of membrane rafts in subcellular membrane transport, trafficking, and signaling.38,39 Rafts are believed to have ideal features for acting as a sorting mechanism in membrane trafficking due to their proposed size and their capacity to sequester both lipids and proteins.38 In our candidate lipid raft protein list, membrane trafficking–related processes were significantly enriched based on DAVID bioinformatic analysis. For example, significantly enriched biological processes included protein transport, vesicle-mediated transport, and Golgi vesicle transport. The pathway of SNARE interactions in vesicular transport was also significantly enriched. Proteins detected in the DRM fraction of lens fiber cells that are involved in SNARE interactions included Vesicle-trafficking protein SEC22b (SEC22b), Synaptobrevin homolog YKT6 (YKT6), Synaptosomal-associated protein (SNAP23), Vesicle transport through interaction with t-SNAREs homolog 1B (VTI1B), Vesicle transport protein USE1 (USE1), several vesicle-associated membrane proteins, and multiple types of syntaxins. Proteins in raft domains that are involved in endocytic trafficking37 are also detected, including caveolin-1, Cell division control protein 42 homolog, Transforming protein RhoA, ADP-ribosylation factor 1, and flotillins. These results strongly support the concept that lipid raft domains play an important role in subcellular trafficking within lens fiber cells. The most important role of rafts at the cell surface may be their function in signal transduction and the small GTPases are central to many signaling processes.40 In our list of proteins detected in the DRM fractions, enriched biological processes include small GTPase-mediated signal transduction. In addition, organization and clustering of lipid rafts into more active signaling platforms depends on interaction with and dynamic rearrangement of the cytoskeleton41 and many of the structural and functional properties of rafts require an intact actin cytoskeleton.44 Cytoskeleton organization was one of the enriched biological processes and regulation of actin cytoskeleton was one of the enriched pathways.
In conclusion, we have identified 506 proteins in high-buoyancy DRM fractions of lens fiber cell membranes and more than 85% of identified proteins have been reported to have lipid raft localization in other cells. The iTRAQ analysis of the high-buoyancy DRM proteins showed variable cholesterol removal dependencies suggesting the presence of true lipid raft proteins and some lipid raft–associated proteins. Enriched GO categories and pathways strongly support that lipid raft domains play important roles in the lens; roles that have not been widely explored. Considering the important functions of lipid raft domains in other cell types, including roles in trafficking, signal transduction, apoptosis, and cytoskeletal organization, it will be interesting to study the role of lens fiber cell lipid rafts in differentiation as well as in cell migration and elongation. Considering different lipid composition between human and bovine lens,66 as well as lipid changes with aging,67 further characterization of the lipid raft proteome in the human lens is warranted. It also will be interesting to study whether lipid rafts play a role in lens protein aggregation during lens aging, given their increased affinity for oligomeric proteins, particularly in neurodegenerative diseases.68–70
Supplementary Material
Acknowledgments
Supported by National Institutes of Health Grants EY-13462 (KLS) and P30 EY-008126. The authors acknowledge support from the Vanderbilt University Proteomics Facility in the Mass Spectrometry Research Center.
Disclosure: Z. Wang, None; K.L. Schey, None
References
- 1. Bretscher MS. Membrane structure: some general principles. Science. 1973; 181: 622–629. [DOI] [PubMed] [Google Scholar]
- 2. Simons K,, Ikonen E. Functional rafts in cell membranes. Nature. 1997; 387: 569–572. [DOI] [PubMed] [Google Scholar]
- 3. Pike L. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006; 47: 1597–1598. [DOI] [PubMed] [Google Scholar]
- 4. Brown DA,, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000; 275: 17221–17224. [DOI] [PubMed] [Google Scholar]
- 5. Simons K,, van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988; 27: 6197–6202. [DOI] [PubMed] [Google Scholar]
- 6. Ahmed SN,, Brown DA,, London E. On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble liquid-ordered lipid phase in model membranes. Biochemistry. 1997; 36: 10944–10953. [DOI] [PubMed] [Google Scholar]
- 7. Hanada K,, Nishijima M,, Akamatsu Y,, Pagano RE. Both sphingolipids and cholesterol participate in the detergent insolubility of alkaline phosphatase a glycosylphosphatidylinositol-anchored protein, in mammalian membranes. J Biol Chem. 1995; 270: 6254–6260. [DOI] [PubMed] [Google Scholar]
- 8. Simons K,, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011; 3: a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Byrdwell WC,, Borchman D,, Porter RA,, Taylor KG,, Yappert MC. Separation and characterization of the unknown phospholipid in human lens membranes. Invest Ophthalmol Vis Sci. 1994; 35: 4333–4343. [PubMed] [Google Scholar]
- 10. Zigman S,, Paxhia T,, Marinetti G,, Girsch S. Lipids of human lens fiber cell membranes. Curr Eye Res. 1984; 3: 887–896. [DOI] [PubMed] [Google Scholar]
- 11. Borchman D,, Yappert MC. Lipids and the ocular lens. J Lipid Res. 2010; 51: 2473–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raguz M,, Mainali L,, O'Brien WJ,, Subczynski WK. Lipid-protein interactions in plasma membranes of fiber cells isolated from the human eye lens. Exp Eye Res. 2014; 120: 138–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rujoi M,, Jin J,, Borchman D,, Tang D,, Yappert MC. Isolation and lipid characterization of cholesterol-enriched fractions in cortical and nuclear human lens fibers. Invest Ophthalmol Vis Sci. 2003; 44: 1634–1642. [DOI] [PubMed] [Google Scholar]
- 14. Jacob RF,, Cenedella RJ,, Mason RP. Direct evidence for immiscible cholesterol domains in human ocular lens fiber cell plasma membranes. J Biol Chem. 1999; 274: 31613–31618. [DOI] [PubMed] [Google Scholar]
- 15. Jacob RF,, Cenedella RJ,, Mason RP. Evidence for distinct cholesterol domains in fiber cell membranes from cataractous human lenses. J Biol Chem. 2001; 276: 13573–13578. [DOI] [PubMed] [Google Scholar]
- 16. Cenedella RJ,, Sexton PS,, Brako L,, Lo WK,, Jacob RF. Status of caveolin-1 in various membrane domains of the bovine lens. Exp Eye Res. 2007; 85: 473–481. [DOI] [PubMed] [Google Scholar]
- 17. Tong J,, Briggs MM,, Mlaver D,, Vidal A,, McIntosh TJ. Sorting of lens aquaporins and connexins into raft and nonraft bilayers: role of protein homo-oligomerization. Biophys J. 2009; 97: 2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schroeder R,, London E,, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci U S A. 1994; 91: 12130–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Staneva G,, Seigneuret M,, Koumanov K,, Trugnan G,, Angelova MI. Detergents induce raft-like domains budding and fission from giant unilamellar heterogeneous vesicles: a direct microscopy observation. Chem Phys Lipids. 2005; 136: 55–66. [DOI] [PubMed] [Google Scholar]
- 20. Rappsilber J,, Ishihama Y,, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003; 75: 663–670. [DOI] [PubMed] [Google Scholar]
- 21. Washburn MP,, Wolters D,, Yates JR,, III. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001; 19: 242–247. [DOI] [PubMed] [Google Scholar]
- 22. Dasari S,, Chambers MC,, Slebos RJ,, Zimmerman LJ,, Ham AJ,, Tabb DL. TagRecon: high-throughput mutation identification through sequence tagging. J Proteome Res. 2010; 9: 1716–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma ZQ, Dasari S, Chambers MC, et al. IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009; 8: 3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacLean B,, Tomazela DM,, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010; 26:– 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen X,, Jen A,, Warley A,, Lawrence MJ,, Quinn PJ,, Morris RJ. Isolation at physiological temperature of detergent-resistant membranes with properties expected of lipid rafts: the influence of buffer composition. Biochem. J. 2009; 417: 525–533. [DOI] [PubMed] [Google Scholar]
- 26. Tanner W,, Malinsky J,, Opekarová M. In plant and animal cells detergent-resistant membranes do not define functional membrane rafts. Plant Cell. 2011; 23: 1191–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chamberlain LH. Detergents as tools for the purification and classification of lipid rafts. FEBS Lett. 2004; 559: 1–5. [DOI] [PubMed] [Google Scholar]
- 28. Drevot P,, Langlet C,, Guo XJ,, et al. TCR signal initiation machinery is pre-assembled and activated in a subset of membrane rafts. EMBO J. 2002; 21: 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan Y,, Iwata F,, Wang D,, et al. Identification of aquaporin-5 and lipid rafts in human resting saliva and their release into cevimeline-stimulated saliva. Biochim Biophys Acta. 2009; 1790: 49–56. [DOI] [PubMed] [Google Scholar]
- 30. Ishikawa Y,, Yuan Z,, Inoue N,, et al. Identification of AQP5 in lipid rafts and its translocation to apical membranes by activation of M3 mAChRs in interlobular ducts of rat parotid gland. Am J Physiol Cell Physiol. 2005; 289: C1303–C1311. [DOI] [PubMed] [Google Scholar]
- 31. Ishikawa Y,, Cho G,, Yuan Z,, Inoue N,, Nakae Y. Aquaporin-5 water channel in lipid rafts of rat parotid glands. Biochim Biophys Acta. 2006; 1758: 1053–1060. [DOI] [PubMed] [Google Scholar]
- 32. Liu H,, Sadygov RG,, Yates JR,, III. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004; 76: 4193–4201. [DOI] [PubMed] [Google Scholar]
- 33. Huang DW,, Sherman BT,, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 34. Shah A,, Chen D,, Boda AR,, Foster LJ,, Davis MJ,, Hill MM. RaftProt: mammalian lipid raft proteome database. Nucleic Acids Res. 2015; 43: D335–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poston CN,, Duong E,, Cao Y,, Bazemore-Walker CR. Proteomic analysis of lipid raft-enriched membranes isolated from internal organelles. Biochem Biophys Res Commun. 2011; 415: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bae TJ,, Kim MS,, Kim JW,, et al. Lipid raft proteome reveals ATP synthase complex in the cell surface. Proteomics. 2004; 4: 3536–3548. [DOI] [PubMed] [Google Scholar]
- 37. Browman DT,, Resek ME,, Zajchowski LD,, Robbins SM. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J Cell Sci. 2006; 119: 3149–3160. [DOI] [PubMed] [Google Scholar]
- 38. Diaz-Rohrer B,, Levental KR,, Levental I. Rafting through traffic: membrane domains in cellular logistics. Biochim Biophys Acta. 2014; 1838: 3003–3013. [DOI] [PubMed] [Google Scholar]
- 39. Lingwood D,, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010; 327: 46–50. [DOI] [PubMed] [Google Scholar]
- 40. Simons K,, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000; 1: 31–39. [DOI] [PubMed] [Google Scholar]
- 41. Head BP,, Patel HH,, Insel PA. Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts mediators of cytoskeletal arrangement and cell signaling. Biochim Biophys Acta. 2014; 1838: 532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Causeret M,, Taulet N,, Comunale F,, Favard C,, Gauthier-Rouvière C. N-cadherin association with lipid rafts regulates its dynamic assembly at cell-cell junctions in C2C12 myoblasts. Mol Biol Cell. 2005; 16: 2168–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin D,, Zhou J,, Zelenka PS,, Takemoto DJ. Protein kinase C gamma regulation of gap junction activity through caveolin-1-containing lipid rafts. Invest Ophthalmol Vis Sci. 2003; 44: 5259–5268. [DOI] [PubMed] [Google Scholar]
- 44. Chichili GR,, Rodgers W. Cytoskeleton-membrane interactions in membrane raft structure. Cell Mol Life Sci. 2009; 66: 2319–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li LK,, So L,, Spector A. Membrane cholesterol and phospholipid in consecutive concentric sections of human lenses. J Lipid Res. 1985; 26: 600–609. [PubMed] [Google Scholar]
- 46. Zidovetzki R,, Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim Biophys Acta. 2007; 1768: 1311–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Foster LJ,, De Hoog CL,, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc Natl Acad Sci U S A. 2003; 100: 5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schey KL,, Gutierrez DB,, Wang Z,, Wei J,, Grey AC. Novel fatty acid acylation of lens integral membrane protein aquaporin-0. Biochemistry. 2010; 49: 9858–9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fantini J,, Yahi N. Molecular basis for the glycosphingolipid-binding specificity of α-synuclein: key role of tyrosine 39 in membrane insertion. J Mol Biol. 2011; 408: 654–669. [DOI] [PubMed] [Google Scholar]
- 50. Fantini J,, Barrantes FJ. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front Physiol. 2013; 4: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tong J,, Canty JT,, Briggs MM,, McIntosh TJ. The water permeability of lens aquaporin-0 depends on its lipid bilayer environment. Exp Eye Res. 2013; 113: 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taylor V,, Welcher AA,, Program AE,, Suter U. Epithelial membrane protein-1, peripheral myelin protein 22, and lens membrane protein 20 define a novel gene family. J Biol Chem. 1995; 270: 28824–28833. [DOI] [PubMed] [Google Scholar]
- 53. Maher GJ,, Black GC,, Manson FD. Focus on molecules: lens intrinsic membrane protein (LIM2/MP20). Exp Eye Res. 2012; 103: 115–116. [DOI] [PubMed] [Google Scholar]
- 54. Hasse B,, Bosse F,, Müller HW. Proteins of peripheral myelin are associated with glycosphingolipid/cholesterol-enriched membranes. J Neurosci Res. 2002; 69: 227–232. [DOI] [PubMed] [Google Scholar]
- 55. Lee S,, Amici S,, Tavori H,, et al. PMP22 is critical for actin-mediated cellular functions and for establishing lipid rafts. J Neurosci. 2014; 34: 16140–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gonen T,, Hite RK,, Cheng Y,, Petre BM,, Kistler J,, Walz T. Polymorphic assemblies and crystalline arrays of lens tetraspanin MP20. J Mol Biol. 2008; 376: 380–392. [DOI] [PubMed] [Google Scholar]
- 57. Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005; 6: 801–811. [DOI] [PubMed] [Google Scholar]
- 58. Ren J,, Wen L,, Gao X,, Jin C,, Xue Y,, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel. 2008; 21: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grey AC,, Jacobs MD,, Gonen T,, Kistler J,, Donaldson PJ. Insertion of MP20 into lens fibre cell plasma membranes correlates with the formation of an extracellular diffusion barrier. Exp Eye Res. 2003; 77: 567–574. [DOI] [PubMed] [Google Scholar]
- 60. Biswas SK,, Jiang JX,, Lo WK. Gap junction remodeling associated with cholesterol redistribution during fiber cell maturation in the adult chicken lens. Mol Vis. 2009; 15: 1492–1508. [PMC free article] [PubMed] [Google Scholar]
- 61. Schubert AL,, Schubert W,, Spray DC,, Lisanti MP. Connexin family members target to lipid raft domains and interact with caveolin-1. Biochemistry. 2002; 41: 5754–5764. [DOI] [PubMed] [Google Scholar]
- 62. Lin D,, Lobell S,, Jewell A,, Takemoto DJ. Differential phosphorylation of connexin46 and connexin50 by H2O2 activation of protein kinase Cgamma. Mol Vis. 2004; 10: 688–695. [PubMed] [Google Scholar]
- 63. Ifeanyi F,, Takemoto L. Differential binding of alpha-crystallins to bovine lens membrane. Exp Eye Res. 1989; 49: 143–147. [DOI] [PubMed] [Google Scholar]
- 64. Borchman D,, Tang D. Binding capacity of alpha-crystallin to bovine lens lipids. Exp Eye Res. 1996; 63: 407–410. [DOI] [PubMed] [Google Scholar]
- 65. Mulders JW,, Stokkermans J,, Leunissen JA,, Benedetti EL,, Bloemendal H,, de Jong WW. Interaction of alpha-crystallin with lens plasma membranes. Affinity for MP26. Eur J Biochem. 1985; 152: 721–728. [DOI] [PubMed] [Google Scholar]
- 66. Deeley JM,, Mitchell TW,, Wei X,, et al. Human lens lipids differ markedly from those of commonly used experimental animals. Biochim Biophys Acta. 2008; 1781: 288–298. [DOI] [PubMed] [Google Scholar]
- 67. Hughes JR,, Deeley JM,, Blanksby SJ,, et al. Instability of the cellular lipidome with age. Age. 2012; 34: 935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Paladino S,, Sarnataro D,, Pillich R,, Tivodar S,, Nitsch L,, Zurzolo C. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J Cell Biol. 2004; 167: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ikeda K,, Yamaguchi T,, Fukunaga S,, Hoshino M,, Matsuzaki K. Mechanism of amyloid β-protein aggregation mediated by GM1 ganglioside clusters. Biochemistry. 2011; 50: 6433–6440. [DOI] [PubMed] [Google Scholar]
- 70. Sonnino S,, Aureli M,, Grassi S,, Mauri L,, Prioni S,, Prinetti A. Lipid rafts in neurodegeneration and neuroprotection. Mol Neurobiol. 2014; 50: 130–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.