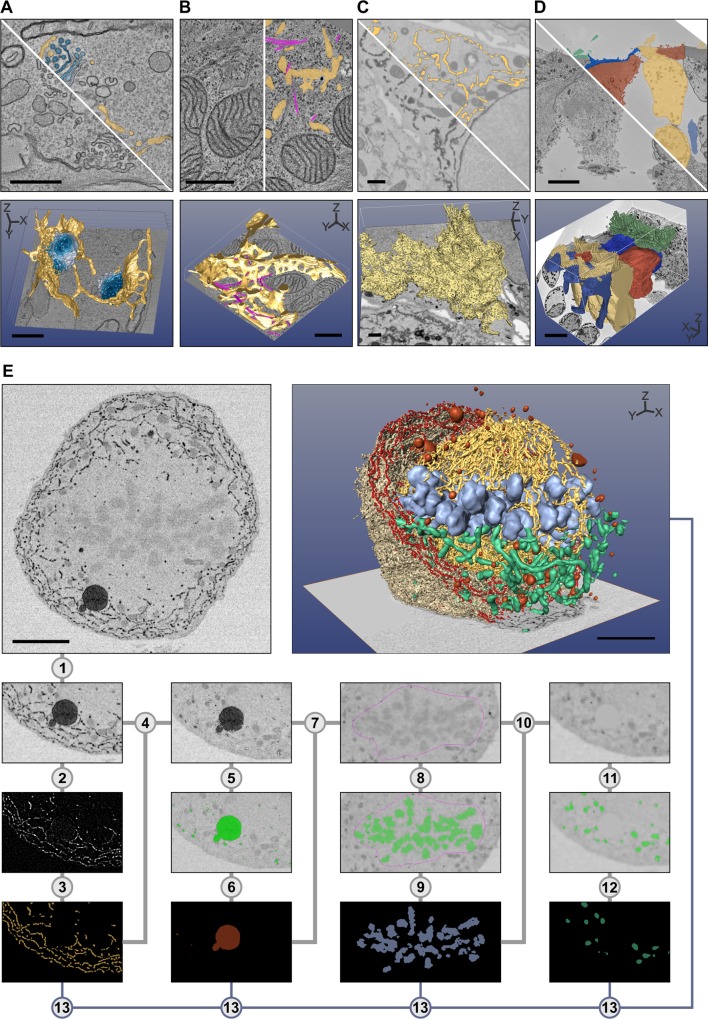
Fig 2. The selection of the best suitable tool for segmentation depends on specimen and object of interest.
Five examples are given: in (A–D), the top row of each image shows a preprocessed slice and a segmentation overlay, and the bottom row shows the final 3-D visualization. Segmentation was done with MIB, and the 3-D rendering using different freeware (A–D) and commercial (E) software packages. (A) Trypanosoma brucei was chemically fixed and imaged with electron tomography (ET). Each Golgi cisternae (four shades of blue) was manually segmented using the brush tool, while ER and ER-derived vesicles (yellow) were segmented using a combination of the brush tool and shape interpolation. The resulting 3-D model was rendered directly in MIB. (B) A Huh-7 cell was high-pressure frozen and freeze-substituted, and a portion of the cell was subjected to ET [11]. ER (yellow) was segmented using the brush tool with shape interpolation and microtubules (magenta) using the line tracker tool. The resulting model was exported in the IMOD-compatible format and rendered in IMOD [6]. (C) A Huh-7 cell transiently expressing ssHRP-KDEL was cytochemically stained (dark precipitate) and imaged with a serial block-face scanning electron microscope (SB-EM) [11]. The ER network (yellow) was segmented semiautomatically using global black-and-white thresholding and further polished using quantification filtering [11]. The model was exported in the nearly raw raster data (NRRD) format and rendered in 3D Slicer [8]. (D) Mouse cochlea was perilymphatically fixed, and the sensory epithelium of the medial part of the cochlear duct was imaged with SB-EM [13]. Different cell types of the organ of Corti (inner hairs in green, outer hair cells in yellow, external rod in vermilion, internal rod in sapphire blue, and phalangeal part of the Deiters’ cells in greyish blue) were segmented using local thresholding combined with shape interpolation and model rendered using 3D Slicer. (E) Stepwise segmentation workflow is needed to generate a 3-D model of a complex structure. A metaphase Huh-7 cell transiently expressing ssHRP-KDEL was cytochemically stained (dark precipitate in ER lumen) and imaged with SB-EM [10]. The modelling workflow for ER consists of 13 steps (S1 Table). For the visualization, the model was exported in the AmiraMesh format and rendered in Amira. Scale bars: A, B 500 nm; C, D, E 5 μm.