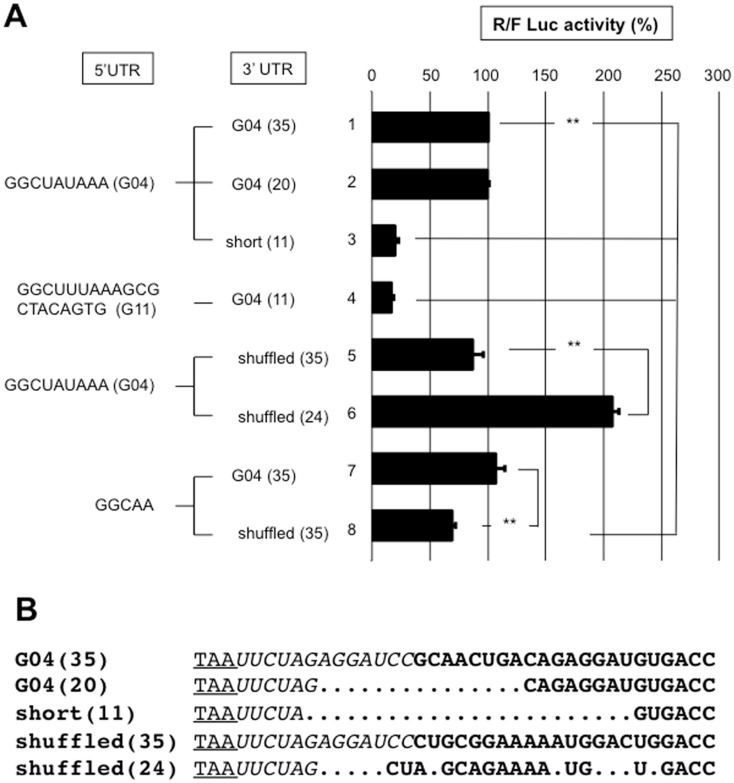
Fig 9. The roles of the 3'UTRs sequences on NSP3-dependent translation efficiency.
A: BSRT7 cells expressing NSP3 were electroporated with the S-RNA and capped R-RNA with the 5'UTR from gene 4 (lines1-3,5,6), 11 (line 4) or a 5 nt-long UTR (lines 7,8) combined with the full-length 3’UTR from gene 4 wild-type (lines 1,4,7); shuffled (lines 5,8); shortened to 20 (line 2) or 11 (lines 3,4) nt; or shortened to 24 nt and shuffled (line 6). The numbers in the parenthesis indicate the 3'UTR length. B: An alignment for the 3’UTR is shown, wherein the stop codon is underlined, the nucleotides transcribed from the vector are italicized, and the nucleotides from the gene 4 3'UTR are in bold. The alignment was generated manually. The Renilla and firefly luciferases activities were measured 6 hours after electroporation. The ratio of Renilla to firefly luciferase activities is reported relative to the control (line 1), which was considered 100. The data are the mean ± SEM of three independent experiments in triplicate. A Student’s t-test was used (*p<0.05;**p<0,01).