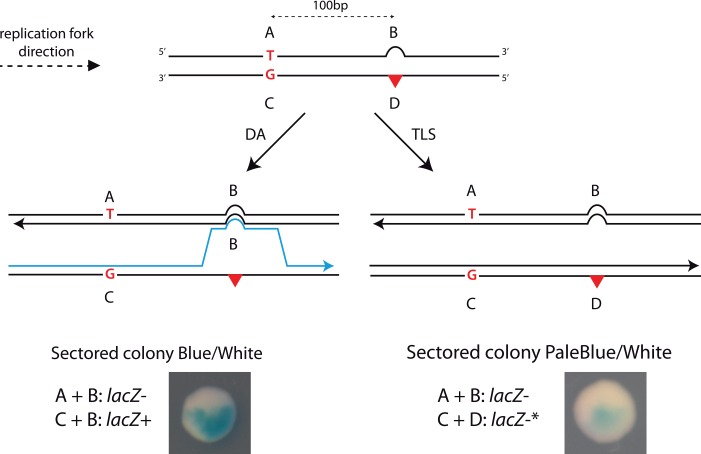
Fig 1. Genetic system to monitor HDGR in the presence of different DNA lesions.
The system is a modified version of a previous construction used to specifically monitor TLS events [17]. The scheme represents the situation in which the lesion (red triangle) is located in the 5'-end of the lacZ gene in the leading orientation. The damaged strand containing the marker D, where the lesion is located, and the marker C, placed 100 bp upstream the lesion, contains a +2 frameshift in order to inactivate the lacZ gene. Opposite the marker D we introduced a +4 loop (marker B) that restores the reading frame of lacZ, and in the same strand we added marker A that contains a stop codon. Therefore the two strands are lacZ-. Only a mechanism of HDGR by which replication has been initiated on the damaged strand (incorporation of marker C), and where a template switch occurred at the lesion site (leading to incorporation of marker B) will restore a lacZ+ gene (the combination of markers C and B contains neither a stop codon nor a frameshift). *For the combination of marker C and D, we observed a leaky activity of the β-Galactosidase (due to a translational frameshift) giving rise to pale blue colonies.