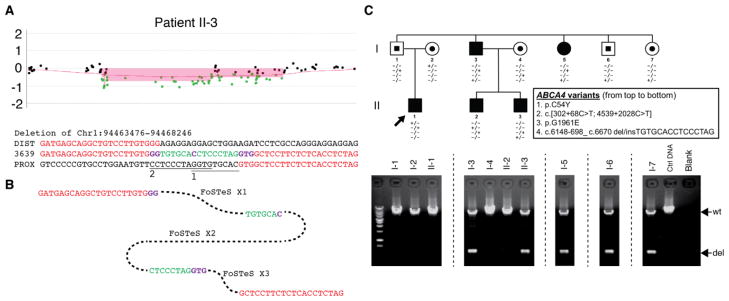
Figure 5.
The deletion/insertion variant in the ABCA4 locus in the family. A. aCGH log2 ratio plot showing the heterozygous deletion identified in patient II-3. The breakpoint junction sequences alignment underneath the plot reveals two inserted DNA fragments (green) conjugated by microhomologies (purple). The potential origins of the inserted DNA fragments are marked with underlines labeled with ”1” and “2”, respectively. DIST/PROX, human genome reference sequences at the distal/proximal end of the junction. B. Potential molecular mechanism explaining the deletion etiology. FoSTeS, fork stalling and template switching. C. Pedigree of the family identified with the deletion/insertion variant allele (c.6148-698_c.6670del/insTGTGCACCTCCCTAG). The gel pictures underneath the pedigree demonstrate the segregation of the deletion allele with three other potential disease-causing alleles identified in this family. wt, wild type allele; del, deletion/insertion variant allele.