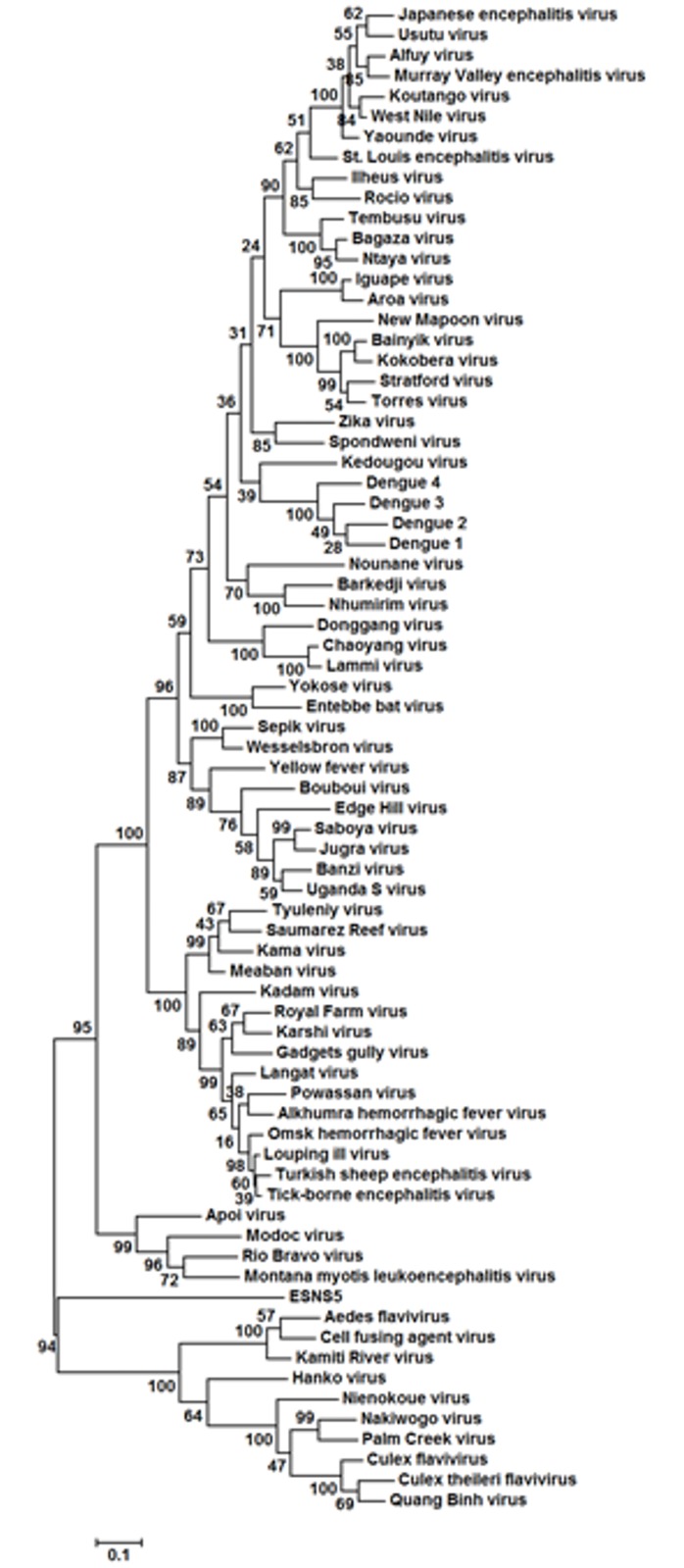
Fig 2. Phylogenetic position of ESNS5.
The evolutionary history of ESNS5 and almost all known flaviviruses was inferred by generating homologous NS5 amino acid sequences using MUSCLE multiple sequence alignment and Gblocks 0.91b software, followed by the Maximum Likelihood methods based on the JTT matrix-based models. The tree with the highest log likelihood (-27149.5525) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial trees for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 74 sequences. All positions containing gaps and missing data were eliminated. There were a total of 496 positions in the final dataset. Evolutionary analysis was conducted in MEGA6.