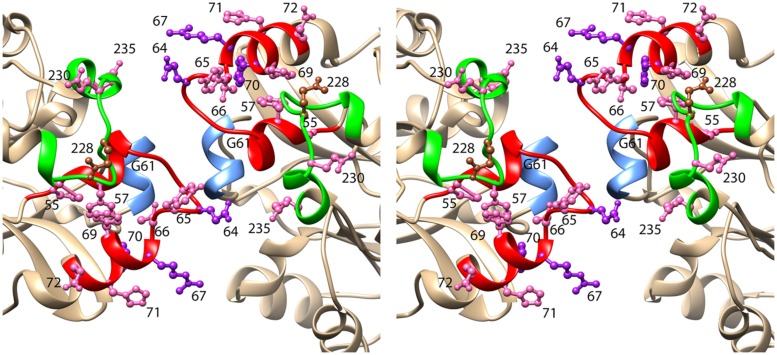
FIGURE 4.
Stereoview of the model of CynD based on the structure of the oxy-nitrilase from Synechocystis sp. PCC6803 (PDB id: 3WUY). The ribbon depicting region 1 is colored red. This region includes three residues that when mutated to cysteine, caused CynD to lose activity: E64, R67, and Y70 (colored purple). It also included seven residues that when mutated to cysteine caused the activity to drop below 50% of the “wt” activity: P55, F57, Y65, T66, F69, H71, and E72 (colored pink). The ribbon depicting region 2 is colored green. It includes two residues that when mutated to cysteine caused the activity to drop below 50% of the “wt” activity: N230 and E235 (colored pink). Residue Q228, mutation of which caused aggregation of the short spirals without loss of activity is colored brown.