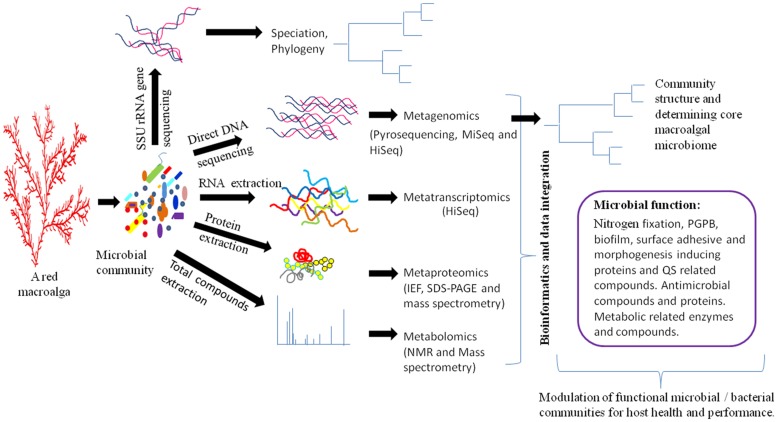
FIGURE 1.
Schematic diagram for elucidating the functions of the macroalgal microbiome. Different meta-omics techniques can be used to identify active functional microbial/bacterial communities. Subsequently, these communities can be used to modulate macroalgal growth and health. Metagenomics and metatranscriptomics analyses can be obtained by Mi-Seq, Hi-Seq, 454 GS FLX SOLiDv4, and Sanger 3730xl sequencing methods (Liu et al., 2012). Functional annotation of metagenomic data can be determined by MG-RAST, IMG/M, METAREP, CAMERA, and MEGAN4 softwares (Kim et al., 2013). Metaproteomics of microbial data can be analyzed by isoelectric focusing (IEF), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS), and liquid chromatography–mass spectrometry (LC-MS/MS) (Kan et al., 2005). The metabolome of microbial communities can be identified by nuclear magnetic resonance spectroscopy and diverse mass spectrometry methods including MALDI-TOF and LC-MS/MS (Lee et al., 2012a,b). PGPB- plant growth-producing bacteria, QS- quorum sensing.