Figure 7. Cortical DS and Functional Connectivity.
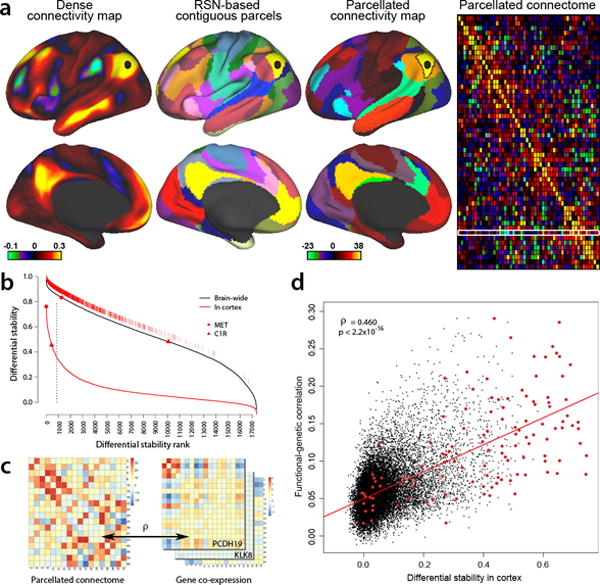
(a) Resting-state functional connectivity from Human Connectome Project (HCP) data (www.humanconnectome.org;29, 45). (Column 1): Functional connectivity from a seed in lateral parietal cortex (black disk), based on a group-average of 468 HCP subjects and showing the full correlation results (scale: Fisher z-transformed ρ); dataset accessible via https://db.humanconnectome.org/data/projects/HCP_500). (Column 2): 52 left-hemisphere contiguous parcels (50 mm2 or larger) from the 17 resting-state networks (RSN) identified by Yeo et al. (2011). (Column 3): parcellated connectivity map for a default-mode network (DMN) parcel (black outline) containing the selected seed, based on 447 HCP subjects (a subset of the above 468) and using partial correlation (scale: z-score). (Column 4): Group-average (n=447) parcellated connectome showing relative connection strength between regions, also based on partial correlation. White rectangular outline identifies connectivity map shown in col. 3. (b) Ranked cortical DS genes (solid red) and brain wide DS genes (black). Top 5th percentile g=867 cortex DS genes (vertical line) are shown as hatch marks just above the whole-brain DS curve. Red disk and triangle show two genes with differing brain-wide and cortical DS. (c) Functional correlation of parcellated RSN connectome (left panel) is compared with genetic co-expression similarity for each gene in each subject (right panel), using the same set of cortical parcels and by calculating the Pearson’s correlation between the vectorized upper diagonal elements of the matrices. (d) DS versus functional-genetic correlation for 17,348 genes. Higher cortical DS genes are more predictive of functional cortical connectivity (ρ=0.46, p<2.2e-16), whereas the correlation is substantially weaker for brain wide DS (ρ=0.17; data not shown). Red points are 132 genes identified as drivers of functional connectivity in a postmortem brain tissue data set whose polymorphisms significantly affect resting-state functional connectivity in a large sample of healthy adolescents28.
