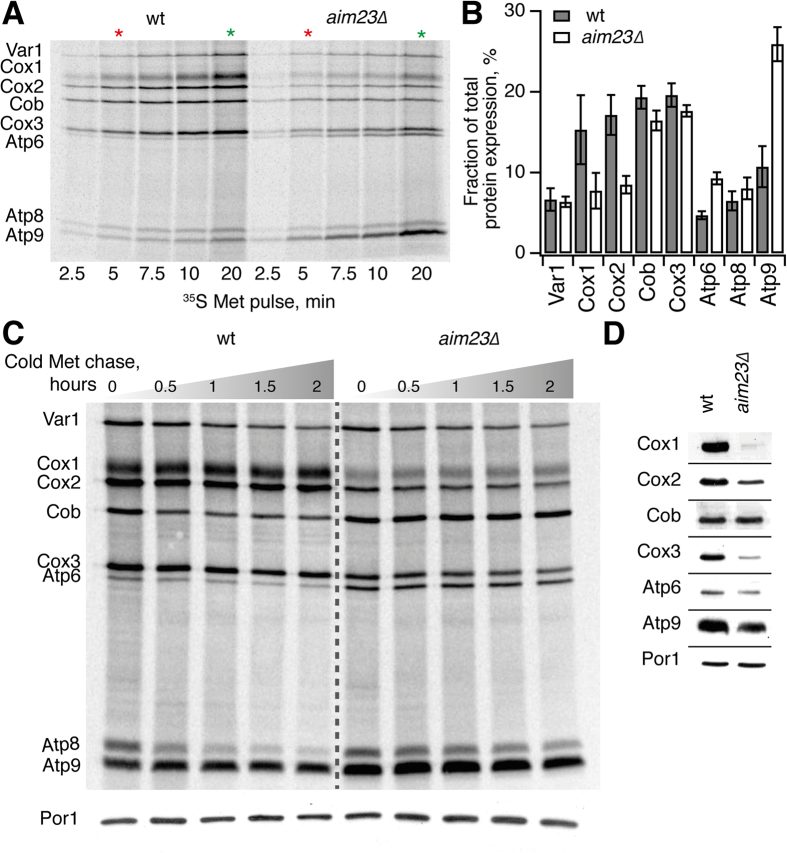
Figure 3. Lack of mIF3/AIM23 leads to unbalanced synthesis of proteins encoded in mtDNA.
The experiments were performed using a congenic set of wild type and aim23∆ strains in D273-10B background. (A) Time course of 35S-methionine incorporation in mitochondrially synthesized proteins in live yeast cells. Cytoplasmic translation was suppressed by the addition of 0.2 mg/ml cycloheximide as per Gouget and colleagues32. 5 minutes (red asterisk) and 20 minutes (green asterisk) time points were used for quantitative analysis of relative protein expression presented on Fig. 3B and Supplementary Figure 2, respectively. (B) Levels of mitochondrially-encoded proteins after 5 min labeling with 35S-methionine. The relative expression is normalized to total expression of mitochondrially encoded protein genes. Error bars indicate the standard deviation of the mean of at least three independent experiments.(C) Turnover of mitochondrially synthesized proteins in wild type and aim23∆ strains. After 15 minutes of 35S methionine pulse labeling was carried out as per Gouget and colleagues32, the labeling reaction was stopped by the addition of cold methionine (final concentration of 80 mM) and puromycin (final concentration of 4 μg/ml). Samples were collected after the indicated time points, proteins were resolved on SDS PAGE and visualized by radioautography. Western blot detection of Porin 1 (Por1) was used as a control for equal loading. Two additional biological replicates of the experiment are presented as a Supplementary Figure 3. (D) Western blot analysis of steady-state levels of mitochondrial proteins in wild-type and aim23∆ strains. Cells were grown until OD600 ≈ 3.0 in liquid YPGal media and mitochondria were isolated according to Meisinger and colleagues58. Equal amounts of mitochondrial proteins were loaded on SDS-PAGE, transferred to nitrocellulose membrane and detected by immunoblotting.