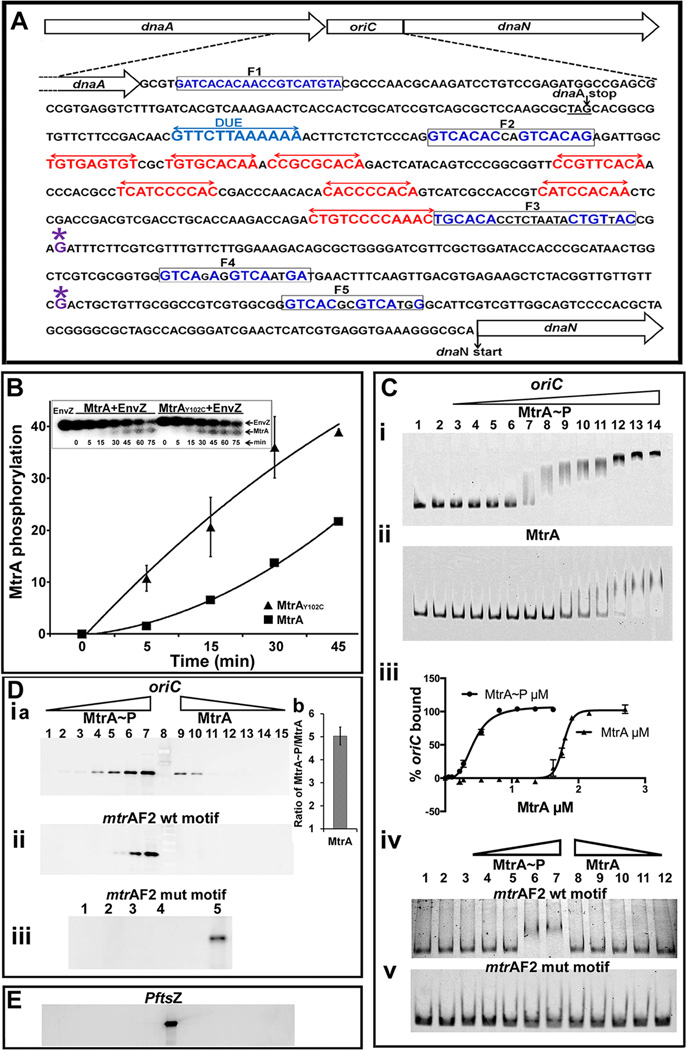
Fig. 1.
(A) M. tuberculosis oriC region and oriC sequence with DnaA- and MtrA- boxes. The dnaA-dnaN intergenic region of M. tuberculosis containing DnaA-boxes (red double arrows), MtrA-motifs (boxed) and DUE sequence (blue double arrows) is shown. For clarity, only the DnaA-boxes defined based on our earlier DMS footprinting data are shown (Madiraju et al., 2006). Although not shown, DNaseI footprinting data located two additional boxes in the 3’ end of oriC, presumably these are low-affinity DnaA- boxes (Zawilak et al., 2004). Presumptive transcription start sites at −110 (downstream of F4 box) and −258 (downstream of F3 box) relative to dnaN start codon based on M. bovis BCG sequence are marked with ‘*’ above the ‘G’ residue (Salazar et al., 2003). (B) Kinetics of MtrA phosphorylation. Autophosphorylated EnvZ was incubated with MtrA or MtrAY102C, samples at indicated time periods were removed, resolved by SDS-PAGE, autoradiography was performed, signals were quantitated by densitometry on a Bio-Rad Molecular imager and data plotted. Inset shows a representative SDS-PA gel autoradiograph image of radiolabeled MtrA and EnvZ proteins. (C) EMSA showing MtrA binding to oriC (panels i, ii) and DNA fragments containing MtrA F2 WT and mutant sequences (panels iv, v). Lanes 1 and 2 (panels i– v) are controls wherein DNA alone (lane 1) or that incubated with EnvZ in the absence of MtrA (lane 2) were resolved. MtrA-mCherry~P was used at 0.054, 0.108, 0.216, 0.27, 0.54, 0.81, 1.35, 1.62, 1.89, 2.16 and 2.7 µM (panels C-i) and MtrA-mCherry at 0.216, 0.27, 0.54, 0.81, 1.08, 1.35, 1.62, 1.75, 1.8, 1.9, 2.16 and 2.7 µM (panels C-ii). Binding data were quantified and the percent oriC bound was calculated (panel iii). MtrA-binding to F2 WT (panel iv) and mutant sequence (panel v) were performed at 0.27, 0.54, 1.08, 2.16 and 2.7 µM protein concentration. (D) Solid phase DNA-binding assays. Streptavidin magnetic beads conjugated with biotinylated oriC (panel i) or MtrA F2 box sequences (panels ii, iii) were incubated with MtrA and processed as described in methods section. MtrA proteins were used at 0.0365, 0.0735, 0.147, 0.294, 0.588, 1.176, 2.32 µM. MtrA~P or MtrA bound to oriC was determined by densitometry and MtrA~P/MtrA calculated for 1.176, 2.32 µM (Panel D-i b). Binding experiments with MtrA-F2 mutant box sequence were performed in duplicate at 1.176 µM (panel D-iii, lanes 1 and 2) and 2.32 µM MtrA (panel D-iii, lanes 3 and 4) respectively, as other concentrations did not show any binding. (E) Binding assays were performed with PftsZ and immunoblotted with α-MtrA as described above. All binding conditions were similar to oriC (D-i). Note: Lanes ‘8’ in D-i a and E and ‘5’ in D-iii are positive controls wherein streptavidin beads conjugated to oriC or MtrA F2 box, respectively, were incubated with 1. 176 µM MtrA~P.