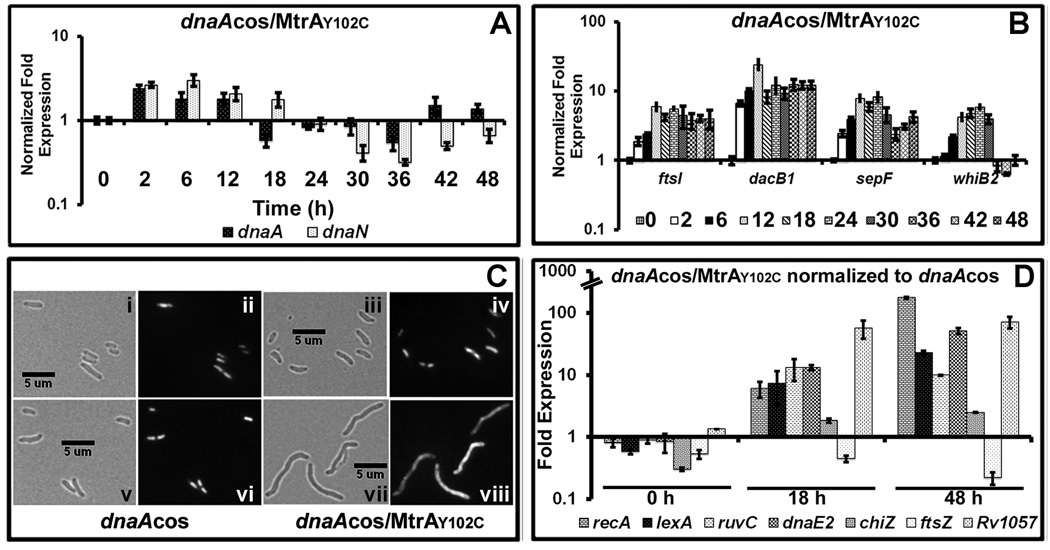
Fig. 7.
Characterization of dnaAcos/MtrAY102C: (A) qRT-PCR analysis of select MtrA targets. Total RNA was extracted from dnaAcos/mtrAY102C at indicated time points and fold expression relative to 16S rRNA was calculated as described in Fig. 4A. (B) qRT-PCR analysis of select cell-division targets ftsI, dacB1, sepF and whiB2 relative to 16S rRNA. (C) Cell morphology of dnaAcos and dnaAcos/MtrAY102C: The dnaAcos (panels i, ii, v, vi) and dnaAcos/MtrAY102C (panels iii, iv, vii, viii) cultures were visualized by microscopy at 0 h (i– iv) and 48 (v– viii) h after initiation of synchronous replication. Brightfield (i, iii, v, vii) and respective fluorescence images obtained following propidium iodide staining (ii, iv, vi, viii) are shown. (D) qRT-PCR analysis of SOS target gene expression. Total RNA from synchronously replicating cultures of M. tuberculosis dnaAcos and dnaAcos/MtrAY102C at indicated time points was extracted and the expression levels of recA, lexA, ruvC, dnaE2, chiZ ftsZ and Rv1057 were determined. Data were normalized to 16SrRNA and fold expression relative to 30°C was determined. Final fold expression was calculated by normalizing dnaAcos/MtrAY102C expression at 0, 18 and 48 h to the respective time points of dnaAcos.