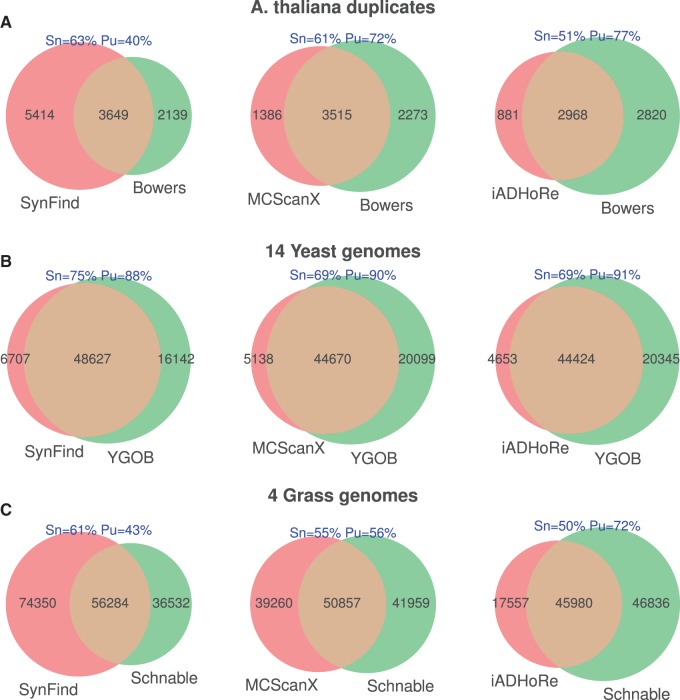
Fig. 5.—
Comparison of SynFind, MCScanX, and iADHoRe on curated data sets. (A) Arabidopsis thaliana alpha, beta, and gamma duplicates from Bowers et al. (2003). (B) Yeast genomes from YGOB (Byrne and Wolfe 2005). (C) Grass genomes from Schnable et al. (2012). Sn: sensitivity, defined as common items divided by total items in truth set; Pu: Purity, defined as common items divided by total items in the test set.