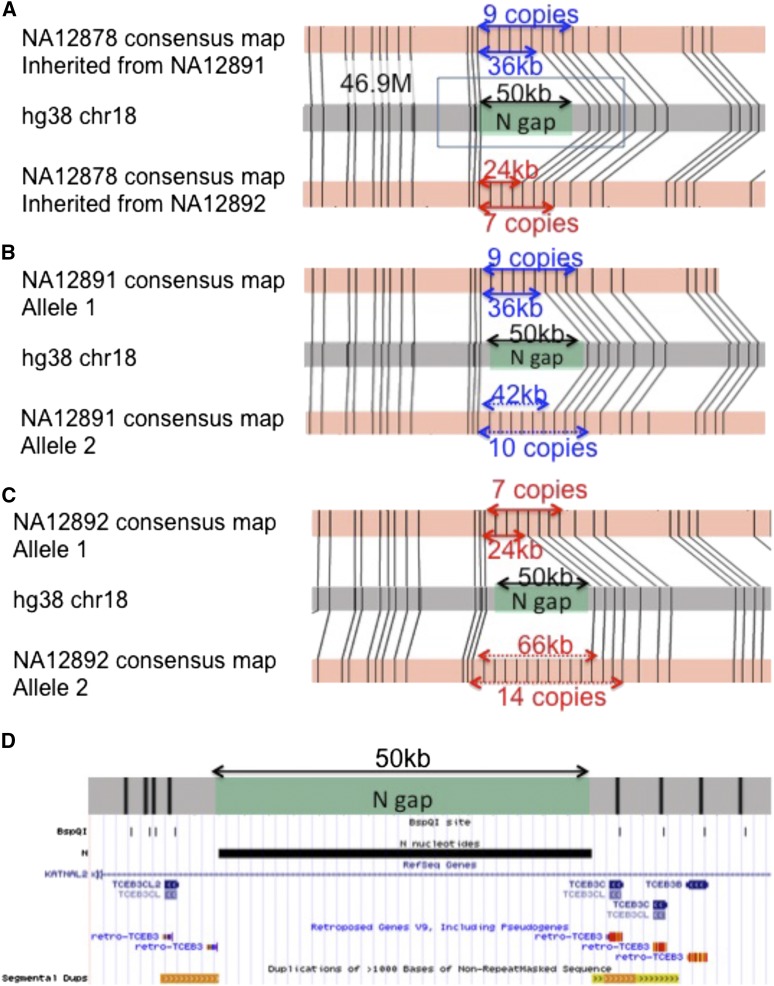
Figure 4.
Use of genome maps to size N‐base gaps and resolve regions with 6-kb tandem repeats. (A) A 50-kb N‐base gap at the chr18:47M region was sized in the trio. NA12878 is heterozygous with 7 and 9 tandem repeats, giving N‐base gap sizes of 24 kb (red arrows, inherited from mother, NA12892) and 36 kb (blue arrows, inherited from father, NA12891). (B) NA12891 has 9 and 10 tandem repeats, giving N‐base gap sizes of 36 kb (blue arrows) and 42 kb (blue dotted arrows). (C) NA12892 has 7 and 14 tandem repeats, giving N‐base gap sizes of 24 kb (red arrows) and 66 kb (red dotted arrows). (D) UCSC genome browser view of the region marked by black box in A. In silico BspQI map shown by the BspQI track overlapped with TCEB genes. The variable number of repeats in this region may thus reflect a copy number difference of the TCEB3 family of genes.