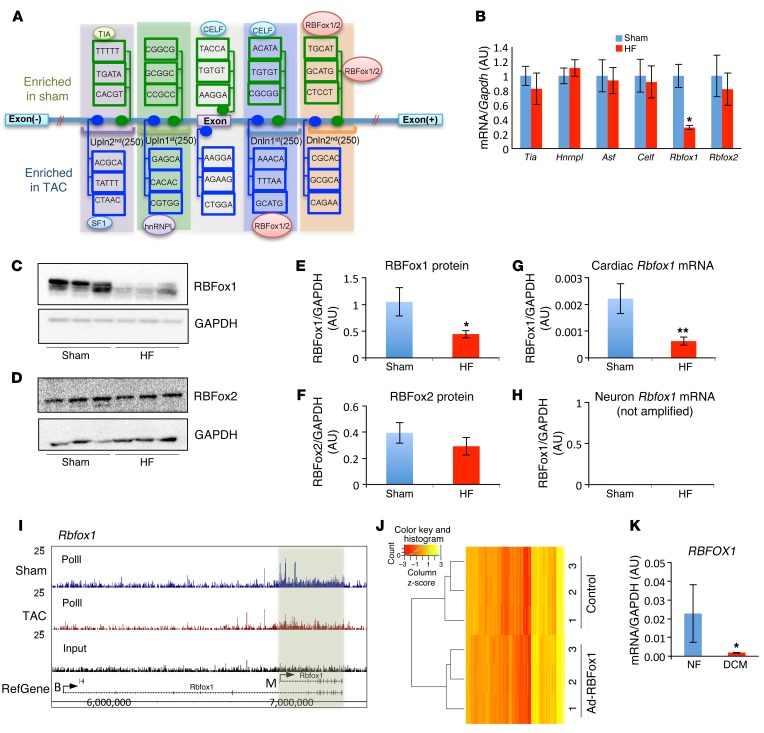
Figure 1. RBFox1 is a key splicing regulator repressed in failing hearts.
(A) Schematic of de novo motif discovery from alternative splicing events. A total of five regions — exon; upstream first 250 bp (Upln 1st); upstream second 250 bp (Upln 2nd); downstream first 250 bp (Dnln 1st); and downstream second 250 bp (Dnln 2nd) — were analyzed. The enriched and conserved motif were indicated with their binding protein. (B) Relative mRNA levels of 6 enriched splicing regulators in sham hearts and hearts after TAC (HF) (n = 3 from each group). Data in both C and D were normalized to GAPDH. Western Blot analysis of RBFox1 and RBFox2 expression levels in normal (Sham) and TAC-induced failing hearts (HF) (n = 3 each sample). (E and F) Quantification of protein expression levels of (E) RBFox1 and (F) RBFox2 in sham-operated hearts compared with those in TAC-induced failing hearts based on Western blot shown in C and D. (G and H) Real-time PCR was performed in TAC- and sham-operated mouse hearts using primers (see Supplemental Table 3) designed specifically targeting either (G) cardiac or (H) neuron Rbfox1 splicing variants (n = 3 each sample). (I) RNA polymerase II occupation on the mouse Rbfox1 gene in sham-operated hearts compared with that in hearts 4 days after TAC. A fragment density of 25 is shown throughout. (J) Heatmap depicting sample-scaled expression of 132 exons significantly changed in the RBFox1-expressing NRVMs identified by RNA-seq and RASL-seq. The blue line in the key is a histogram of the values plotted in the heatmap. (K) Quantification of RBFOX1 mRNA expression in nonfailing (NF) and dilated cardiomyopathy (DCM) human heart samples (n = 4 from each group). *P < 0.05, **P < 0.01, Student’s t test (B, E, G, and K).