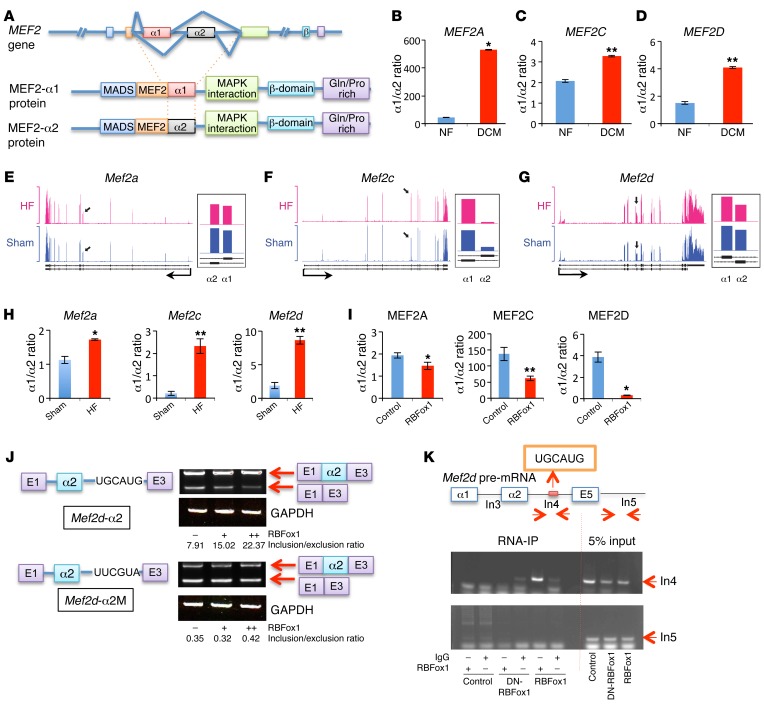
Figure 2. RBFox1 specifically regulates MEF2 α exon inclusion change in failing hearts.
(A) Schematic of the MEF2 gene splicing variants at α exon. (B–D) Quantification of the MEF2 α1/α2 ratio in nonfailing (NF) and dilated cardiomyopathy (DCM) human heart samples (n = 4 from each group). (E–G) Exon-specific RNA reads of the Mef2 genes in failing heart and sham samples based on an RNA-seq data set (43). Black arrows represent the locations of mutually exclusive exons (MXEs) in each gene, and the detail expression profiles (presented as reads per kilobase per million mapped read values) of MXEs at higher magnification are shown to the right. (H) qRT-PCR quantification of Mef2 (Mef2a, Mef2c, and Mef2d) α1 versus α2 transcript ratios from sham hearts (Control) and hearts after TAC (HF) (n = 3 from each group). (I) MEF2 α1/α2 transcript ratio in the control and RBFox1-overexpressing NRVMs (n = 3 from each group). (J) Schematic of the Mef2d α2 exon minigene reporter constructs containing wild-type (Mef2d-α2) or mutated (Mef2d-α2M) RBFox1-binding motif as indicated. Different minigene reporter constructs were transfected alone or in combination with an RBFox1-expressing vector in HEK293 cells. Exon inclusion level was measured by densitometry analysis of RT-PCR products separated by electrophoresis on a 4% agarose gel and indicated as inclusion/exclusion ratio. (K) Cross-link RNA immunoprecipitation assay of Mef2d pre-mRNA sequence. Myoblasts were infected with dominant-negative RBFox1 (DN-RBFox1) and RBFox1 and compared with the mock infected cells. RBFox1 binding to different regions of Mef2d pre-mRNA was detected by semiquantitative RT-PCR as indicated. *P < 0.05, **P < 0.01, Student’s t test (B–D, H, and I).