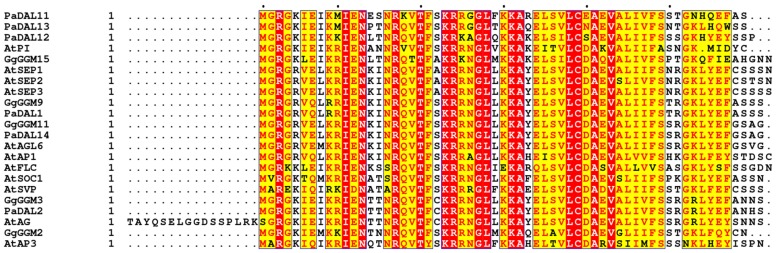
FIGURE 4.
Sequence alignment of MADS TFs M-domain. Aligned M-domain amino acid sequences of A. thaliana SEP3, SEP1, SEP2, AP3, PI, AP1, AG, AGL6, SOC1, SVP, and FLC; the gymnosperm Gnetum gnemon GGM2, GGM15 (AP3/PI-like), GGM3 (AG-like), and GGM9, GGM11 (AGL6-like); and the gymnosperm Picea abies DAL11, DAL12, DAL13 (AP3/PI-like), DAL2 (AG-like), and DAL1, DAL14 (AGL6-like) proteins. Sequence numbering is indicated on the left, with every tenth residue marked by black dots above the sequences. Highly conserved regions are boxed, with similar residues represented in red against a yellow background, invariant residues represented against a red background and non-conserved residues indicated in black.