Figure 1.
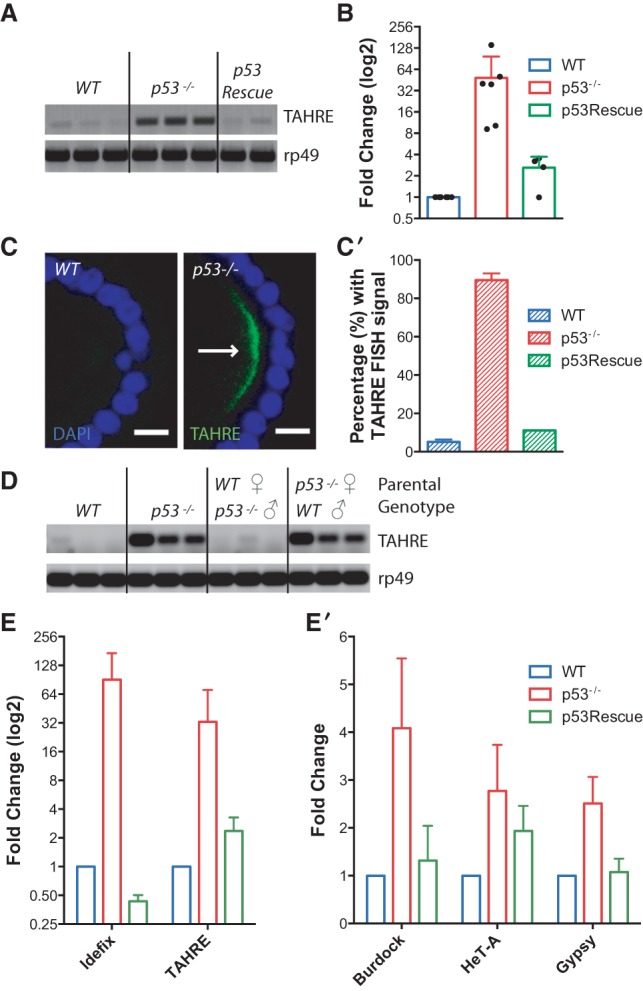
p53 restrains transposon activity in the Drosophila germline. (A) TAHRE retrotransposons, measured by RT–PCR, are highly expressed in dp53− ovaries but minimally expressed in parental wild-type or dp53− flies carrying p53Rescue. The control reference gene ribosomal protein L32 (rp49) is present at similar levels among all genotypes. (B) Derepression of TAHRE transcripts in ovaries of single animals was quantified using ddPCR standardized to the housekeeping gene rp49. Each dot represents measurements from an ovary pair from a single female. TAHRE retrotransposons were consistently dysregulated in dp53− animals (red bar). Normal repression, comparable with wild type (blue bar), occurred when the p53Rescue transgene was present in these mutants (green bar). p53− was significantly different from wild type (P-value = 0.0172) and p53Rescue (P = 0.0347) (see the Materials and Methods). (C,C′) TAHRE expression was assayed by FISH. In C, TAHRE RNAs (arrow) accumulate in the germ plasm of p53− oocytes (stage 9) of stage 9 and 10 egg chambers but not in wild-type egg chambers. (Green) TAHRE signal; (blue) DAPI counterstain. These data are quantified in C′, illustrating TAHRE derepression in p53− ovaries (red bar) Bars, 10 μm. Wild type was significantly different from p53− (P = 0.001) (see the Materials and Methods). (D) TAHRE transcripts, measured by RT–PCR, are maternally loaded into the 1- to 4-h-old embryo. TAHRE elements are derepressed in the p53− embryo (parental genotypes were p53−/−) but undetectable in the wild-type embryo (parental genotypes were wild type). Robust TAHRE expression was also observed in embryos from p53− mothers mated to wild-type fathers (parental genotypes are p53−/− female; wild-type male) but not in embryos from the reciprocal cross (wild-type female; p53−/− male). The control reference transcript rp49 is present at similar levels among all genotypes. Three independent biological replicates are shown for all genotypes. (E,E′) Expression from the indicated retroelements was measured by quantitative RT–PCR. In E the Idefix and TAHRE elements were highly derepressed in dp53− ovaries (red bars) relative to wild-type (blue bars) or p53Rescue (green bars) samples. In E′, retroelements from the Burdock, Gypsy, and HeT-A families (red bars) were similarly but more modestly derepressed in p53− ovaries. Note that in E, the fold change is plotted on a log2 scale to better appreciate differences in transcript levels between wild-type and p53Rescue flies. The error bars represent standard deviations. p53− samples were significantly different from wild type (P-value < 0.05) for Idefix, TAHRE, Burdock, HeT-A, and Gypsy. p53− samples were significantly different from p53Rescue (P -value < 0.05) for TAHRE, Burdock, and Gypsy.
