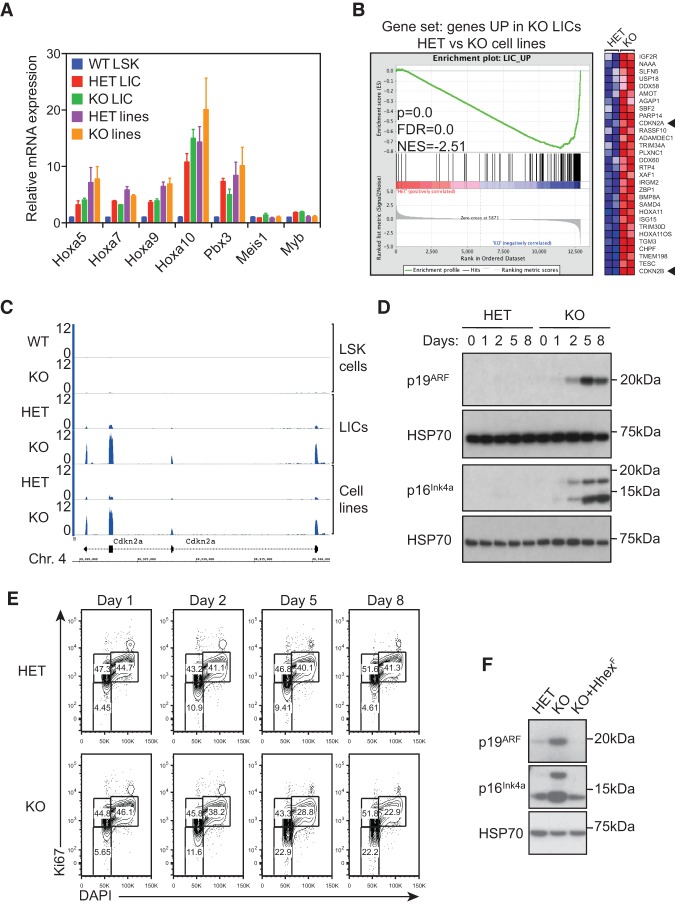
Figure 5.
Up-regulation of p16INK4a and p19ARF tumor suppressor pathways in leukemic cells following Hhex deletion. (A) Maintenance of the MLL-associated self-renewal signature following Hhex deletion. Normalized expression of HoxA and associated genes is shown for wild-type LSK cells and compared with MLL-ENL LICs and cell lines with the indicated Hhex genotype. (B) GSEA showing the association between genes up-regulated following Hhex deletion in LICs and MLL-ENL-induced cell lines. Genes significantly up-regulated in Hhex-deleted LICs were compared in MLL-ENL-induced control (HET) and Hhex-deleted (knockout) cell lines. (Left) In the enrichment plot, skewing to the right indicates that most genes are also up-regulated in Hhex-deleted cell lines. (Right) The heat map shows the top 30 genes most up-regulated in Hhex-deleted cell lines. Cdkn2a and Cdkn2b are indicated by arrowheads. (C) Induction of Cdkn2a expression upon Hhex deletion. Tracks show RNA sequencing coverage of the indicated cell types and Hhex genotypes at the Cdkn2a locus. Units are reads per million mapped reads (RPM). (D,E) Kinetics of p16INK4a and p19ARF expression and cell cycle arrest in Hhex HET and knockout MLL-ENL cell lines following deletion of Hhex. Cells were harvested at the times indicated following tamoxifen (10−9 M) treatment and either lysed in RIPA buffer and processed for immunoblotting with antibodies specific for p16INK4a, p19ARF, and HSP70 (D) or fixed, permeabilized, and stained with DAPI and Ki67-PE-Cy7 for cell cycle analysis by flow cytometry (E). (F) Retroviral transduction of Hhex-null AML cells with Hhex-F suppresses induction of p16INK4a and p19ARF. Hhex HET, Hhex-deleted (knockout), and Hhex-rescued (KO+Hhex-F) cell lines were lysed in RIPA buffer and processed for immunoblotting with antibodies specific for Hhex and as in D.