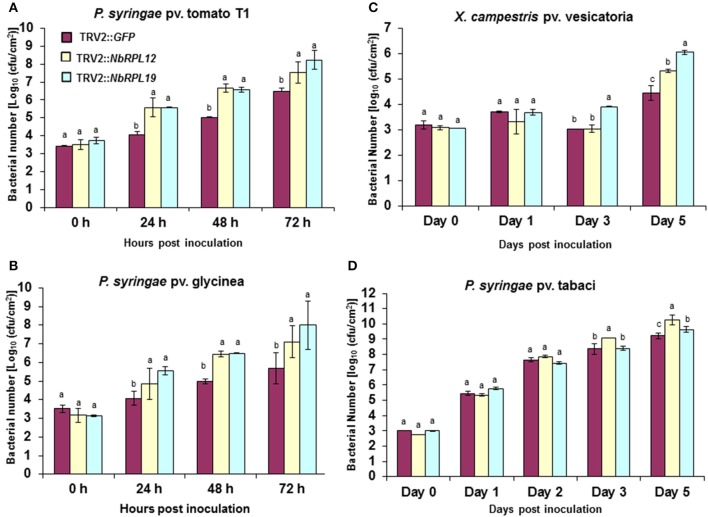
Figure 2.
Multiplication of host and nonhost pathogens in NbRPL12 and NbRPL19 gene silenced N. benthamiana leaves. VIGS was performed using TRV2::NbRPL12 and TRV2::NbRPL19 constructs on 3 week old N. benthamiana plants. Plants inoculated with TRV2::GFP were used as vector control. The silenced leaves were inoculated using needleless syringe with nonhost pathogens, P. syringae pv. tomato T1 (A, OD600 = 0.0002), P. syringae pv. glycinea (B, OD600 = 0.001) and X. campestris pv. vesicatoria (C, OD600 = 0.0002) or host pathogen, P. syringae pv. tabaci (D, OD600 = 0.0001). Bacterial multiplication of host or nonhost pathogens was quantified at various time intervals. The error bars represent standard deviation for three replications. Different letters above the bars indicate a significant difference from Two-way ANOVA at p < 0.05 with Tukey's honest significant differences (HSD) means separation test (α = 0.05) among control and different gene silenced plants within a time point. See Supplementary Table S2 for details about statistics.