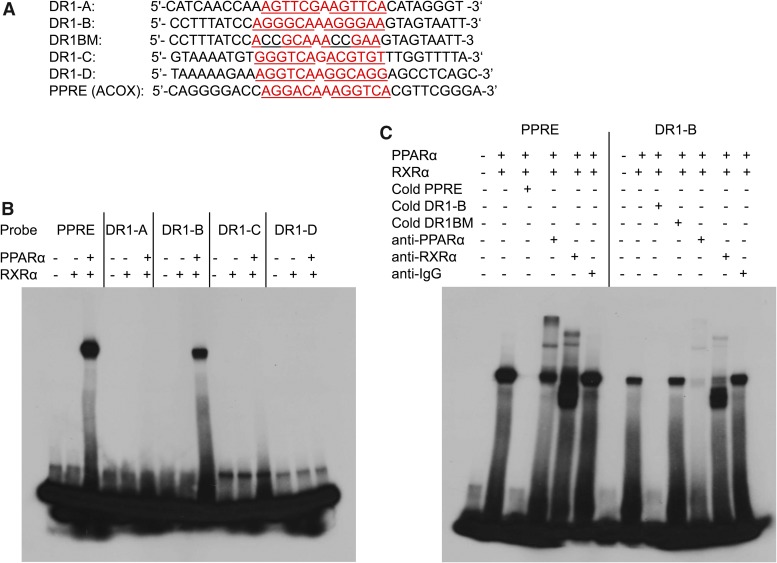
Fig. 6.
Specific binding of PPARα to DR1-B at −2109 bp of the CYP2C8 promoter by gel shift assay. (A) Sequences of the oligonucleotides used for the binding assays. Putative and known PPREs are written in red, and the DR1 half-sites are underlined. GG in DR1-B half-sites were mutated to CC to generate DR1BM. (A) 32P-labeled double-stranded oligonucleotides containing CYP2C8 DR1-A, DR1-B, DR1-C, DR1-D, and a positive control for PPARα binding (PPRE from the rat ACOX1 gene) were incubated for 30 minutes with in vitro transcribed/translated RXRα or hPPARα and RXRα proteins at 4°C. (B) 32P-labeled double-stranded oligonucleotides containing CYP2C8 DR1-B and the known PPRE (rat ACOX1) were incubated for 30 minutes with in vitro translated hPPARα and RXRα at 4°C. In competition experiments, 100-fold excess of unlabeled double-stranded oligonucleotides inhibited formation of the complex. Supershift experiments were performed by incubating the binding reactions with 4 μg of antibodies to PPARα, RXRα, or IgG for 2 hours at 4°C.