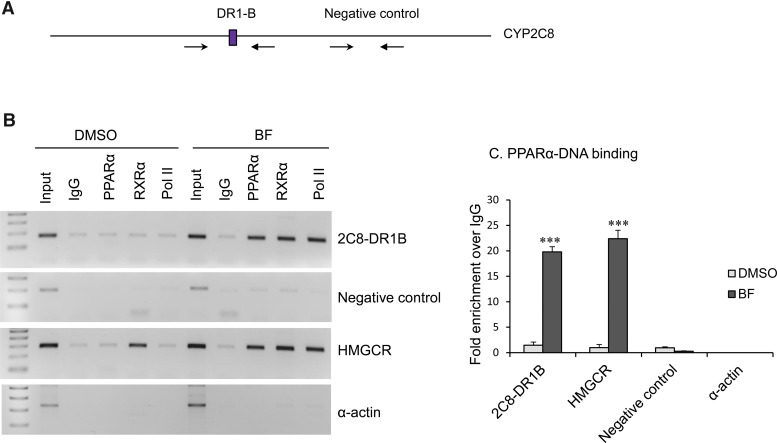
Fig. 7.
ChIP analysis showed in vivo binding of PPARα and RXRα to the CYP2C8 DR1-B sites. (A) CYP2C8 promoter with position of primers used for ChIP PCR analysis. (B) Chromatin was prepared from cultured primary human hepatocytes treated with DMSO control or BF (0.5 mM) for 48 hours, sheared, immunoprecipitated with IgG (negative control), PPARα, RXRα, or RNA Pol II (positive control for transcriptional active genes) antibodies conjugated to Dynabeads protein A/G. PCR was performed with primers spanning the CYP2C8 DR1-B, CYP2C8 promoter region that does not contain DR1 (negative control), PPRE of human HMGCR promoter (positive control for PPARα and RXRα binding), and human α-actin coding region (a negative control for all antibodies). (C) Chromatin extracted from control and BF-treated primary human hepatocytes was immunoprecipitated with antibodies specific to PPARα and IgG. SYBR green qPCR was performed using primers flanking the CYP2C8 DR1-B, CYP2C8 promoter region that does not contain DR1 (negative control), PPRE of human HMGCR, and α-actin. Data represent the human hepatocytes isolated from at least three different donors, as shown in Table 2. Each treatment was analyzed in triplicate, and data indicate the mean ± S.E.M. from at least three different donor preparations. ***P < 0.001 compared with DMSO control (Student’s t test).