Figure 1.
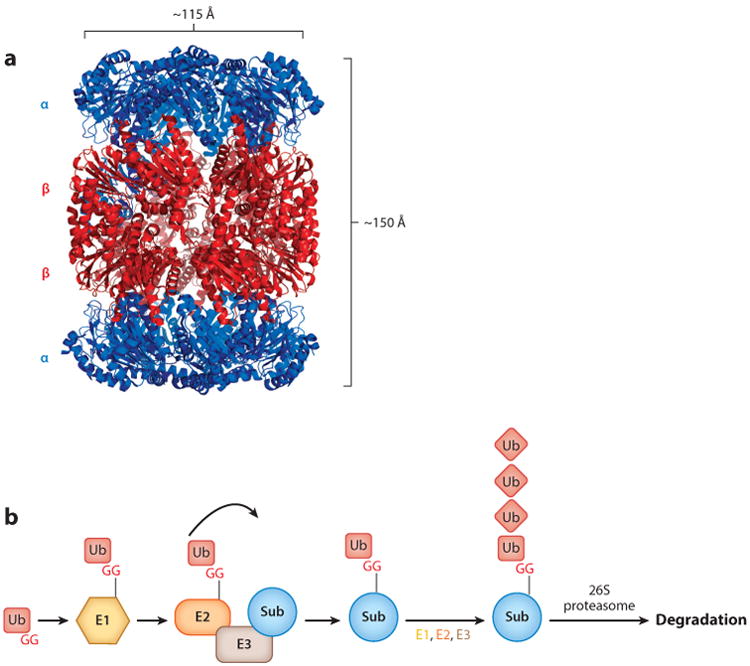
Structure of a proteasome 20S CP and schematic of the Ub-proteasome system. (a) Crystal structure of the 20S CP from Mycobacterium tuberculosis [Protein Data Bank (PDB) ID: 3MI0] (32). (b) Eukaryotic Ub-proteasome system. An E1 enzyme activates Ub by adenylating the C-terminal G of Ub and then forming a thioester bond. Ub is then transferred to an E2 Ub-conjugating enzyme, which then transfers Ub to an E3 ligase that conjugates Ub to a lysine on a target substrate. Additional rounds of ubiquitylation on Ub are generally required for receptors on the 26S proteasome to recognize the doomed protein for degradation. Abbreviations: CP, core particle; G, glycine; Sub, substrate; Ub, ubiquitin.
