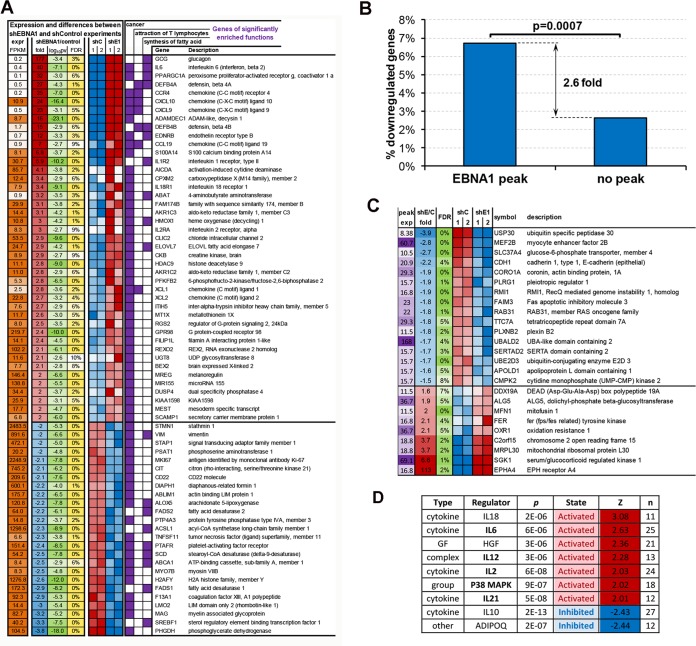
FIG 4.
Integration of RNA-Seq with ChIP-Seq to identify genes regulated directly by EBNA1. LCLs were transduced twice with shControl (shC1 and shC2) or shEBNA1 (shE1 and shE2) and assayed by RNA-Seq. The RNA-Seq data were then analyzed to find significantly affected genes. (A) Genes from 3 major functions significantly affected by EBNA1 depletion. Red, significantly upregulated by EBNA1 depletion; blue, significantly downregulated by EBNA1 depletion. CoA, coenzyme A. (B) Significant association of EBNA1 depletion and downregulation of genes occupied by EBNA1 within 3 kb of the TSS. (C) Genes occupied by EBNA1 within 3 kb of the TSS whose expression levels changed at least 1.5-fold upon EBNA1 depletion. (D) Regulators significantly affected by EBNA1 depletion, as indicated by expression level changes of their known targets. GF, growth factor; HGF, human growth factor; MAPK, mitogen-activated protein kinase.