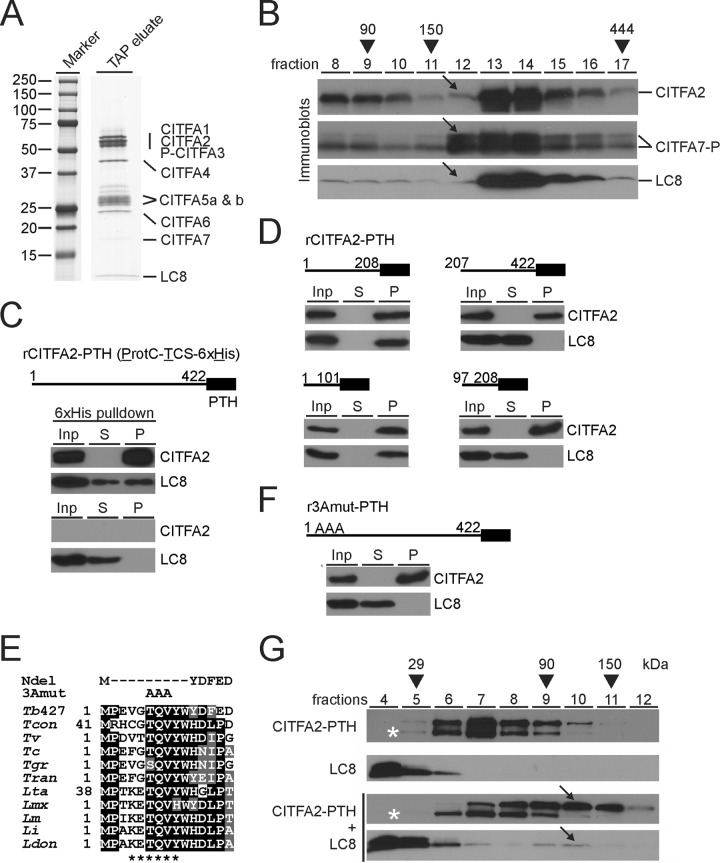
FIG 2.
LC8 binds to the N terminus of CITFA2, promoting its dimerization. (A) The final eluate of a PTP-CITFA3 (P-CITFA3) tandem affinity purification (TAP eluate) was separated on an SDS-polyacrylamide (10 to 20%) gradient gel and stained with Coomassie blue. (B) Sedimentation of tandem affinity-purified CITFA by ultracentrifugation in a 10 to 40% linear sucrose gradient. Fractions, taken from top to bottom, were separated by SDS-PAGE and immunoblotted for specific detection of CITFA2, CITFA7, and LC8. CITFA7-P is so noted due to the presence of ProtC, which remains following tobacco etch virus protease cleavage. Arrows highlight the relative absence of CITFA2 and LC8 in fraction 12. Taq DNA polymerase (95 kDa), IgG (150 kDa), and apoferritin (444 kDa) were analyzed for molecular mass comparison (arrowheads). (C) Pulldown of recombinant, full-length (amino acid residues 1 to 422), wild-type CITFA2 with a C-terminal PTH tag, consisting of ProtC, a thrombin cleavage site (TCS), and a terminal 6×His tag, in the presence of recombinant LC8. Crude extract (input, Inp), supernatant (S), and precipitate (P) were analyzed in relative amounts of 1:1:8 by immunoblotting, using ProtC- and LC8-specific antibodies. A negative-control pulldown assay was conducted in the absence of CITFA2 (bottom panel). (D) Corresponding experiments with CITFA2-PTH fragments which are specified by residue numbers. (E) Alignment of the N-terminal CITFA2 sequences from Trypanosoma brucei brucei strain 427 (Tb427) (accession numbers are listed in Table S1 in the supplemental material), Trypanosoma congolense (Tcon), Trypanosoma vivax (Tv), Trypanosoma cruzi (Tc), Trypanosoma grayi (Tgr), Trypanosoma rangeli (Tran), Leishmania tarentolae (Lta), Leishmania mexicana (Lmx), Leishmania major (Lm), Leishmania infantum (Li), and Leishmania donovani (Ldon). Positions with more than 50% identity or similarity are highlighted in black or gray, respectively. The proposed LC8 binding site is marked below the alignment (asterisks), while the two mutants used for further investigation, NDel and 3Amut, are indicated above. (F) Pulldown assay of full-length rCITFA2-PTH carrying the 3Amut mutation (r3Amut-PTH). (G) Sucrose gradient sedimentation of rCITFA2-PTH alone, LC8 alone, or the two in combination, following coincubation (bottom two panels). Arrows indicate a cosedimentation peak in fraction 10 that was not present when either protein was analyzed on its own. Asterisks identify a truncated form of rCITFA2-PTH.