ABSTRACT
Protease is essential for retroviral replication, and protease inhibitors (PI) are important for treating HIV infection. HIV-2 exhibits intrinsic resistance to most FDA-approved HIV-1 PI, retaining clinically useful susceptibility only to lopinavir, darunavir, and saquinavir. The mechanisms for this resistance are unclear; although HIV-1 and HIV-2 proteases share just 38 to 49% sequence identity, all critical structural features of proteases are conserved. Structural studies have implicated four amino acids in the ligand-binding pocket (positions 32, 47, 76, and 82). We constructed HIV-2ROD9 molecular clones encoding the corresponding wild-type HIV-1 amino acids (I32V, V47I, M76L, and I82V) either individually or together (clone PRΔ4) and compared the phenotypic sensitivities (50% effective concentration [EC50]) of mutant and wild-type viruses to nine FDA-approved PI. Single amino acid replacements I32V, V47I, and M76L increased the susceptibility of HIV-2 to multiple PI, but no single change conferred class-wide sensitivity. In contrast, clone PRΔ4 showed PI susceptibility equivalent to or greater than that of HIV-1 for all PI. We also compared crystallographic structures of wild-type HIV-1 and HIV-2 proteases complexed with amprenavir and darunavir to models of the PRΔ4 enzyme. These models suggest that the amprenavir sensitivity of PRΔ4 is attributable to stabilizing enzyme-inhibitor interactions in the P2 and P2′ pockets of the protease dimer. Together, our results show that the combination of four amino acid changes in HIV-2 protease confer a pattern of PI susceptibility comparable to that of HIV-1, providing a structural rationale for intrinsic HIV-2 PI resistance and resolving long-standing questions regarding the determinants of differential PI susceptibility in HIV-1 and HIV-2.
IMPORTANCE Proteases are essential for retroviral replication, and HIV-1 and HIV-2 proteases share a great deal of structural similarity. However, only three of nine FDA-approved HIV-1 protease inhibitors (PI) are active against HIV-2. The underlying reasons for intrinsic PI resistance in HIV-2 are not known. We examined the contributions of four amino acids in the ligand-binding pocket of the enzyme that differ between HIV-1 and HIV-2 by constructing HIV-2 clones encoding the corresponding HIV-1 amino acids and testing the PI susceptibilities of the resulting viruses. We found that the HIV-2 clone containing all four changes (PRΔ4) was as susceptible as HIV-1 to all nine PI. We also modeled the PRΔ4 enzyme structure and compared it to existing crystallographic structures of HIV-1 and HIV-2 proteases complexed with amprenavir and darunavir. Our findings demonstrate that four positions in the ligand-binding cleft of protease are the primary cause of HIV-2 PI resistance.
INTRODUCTION
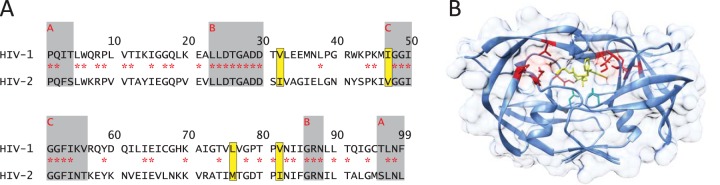
Protease is essential for retroviral replication and is an important target of antiretroviral therapy. In human immunodeficiency virus (HIV), posttranslational processing by the viral protease cleaves the gag and gag-pol polyproteins into their mature functional subunits (1). This process is critical for infectivity and subsequent viral replication. HIV type 1 (HIV-1) and HIV type 2 (HIV-2) proteases are symmetrical homodimers composed of two identical 99-amino-acid subunits, each contributing one aspartic tripeptide motif. Several structural features are conserved between HIV-1 and HIV-2, as well as in proteases from more distantly related retroviruses (2–4). Each monomer includes two layers of orthogonally oriented beta sheets that form a hydrophobic core of the enzyme. Both subunits also contain a flexible beta hairpin known as the “flap region” (residues 42 to 58) that closes down upon binding of a substrate. The active-site aspartates are connected by a network of hydrogen bonds in a structure known as the “fireman's grip” (5), and in both HIV-1 and HIV-2, residues 85 to 88 anchor the catalytic aspartates in the appropriate conformation for substrate cleavage (6–8). In addition, the amino and carboxyl termini of each monomer form a four-stranded beta sheet, which is crucial for dimerization. Alpha-carbon tracings of HIV-1 and HIV-2 structures are almost perfectly superimposable (root mean square [RMS] = 1.0 to 1.1 Å) (9, 10), despite sharing only 38 to 49% identity at the amino acid level (11) (Fig. 1A), and heterodimeric forms of covalently linked HIV-1 and HIV-2 protease monomers are catalytically active (12), providing further evidence for a high degree of structural conservation.
FIG 1.
Sequence and structure of HIV-2 protease. (A) Amino acid alignment of HIV-1NL4-3 and HIV-2ROD9 proteases (adapted from reference 11). Identical residues are marked with red asterisks. Gray boxes labeled A, B, and C indicate the boundaries of the dimerization domain, the active-site/carboxy-terminal triad, and the flap region of protease, respectively. Yellow boxes indicate the residues mutated in this study (amino acid changes I32V, V47I, M76L, and I82V in HIV-2ROD9). (B) Location of the engineered amino acid replacements in the crystal structure of wild-type HIV-2 protease with amprenavir (PDB code 3S45). Active-site aspartate residues D25 and D25′ are shown by green sticks; amprenavir is shown in yellow. Multiple rotamers are shown for M76′ and I32 as deposited in PDB file 3S45.
Despite the structural similarities, HIV-1 and HIV-2 proteases show dramatic disparities in susceptibility to HIV-1 protease inhibitors (PI). HIV-2 is at least partially resistant to the majority of the HIV-1 PI approved by the U.S. Food and Drug Administration (FDA); only lopinavir (LPV), darunavir (DRV), and saquinavir (SQV) are active at clinically useful concentrations (13–20). The determinants of intrinsic PI resistance in HIV-2 are unknown, but previous studies have identified just four residues in the protease binding cleft, at amino acid positions 32, 47, 76, and 82, that differ between HIV-1 and HIV-2 (6, 21–25). Within each virus, the amino acids at these residues are highly conserved in treatment-naive subjects. In HIV-1, a substitution at any one of these residues confers multi-PI resistance (26), and common drug resistance mutations at positions 32, 47, and 82 encode the corresponding wild-type (WT) amino acids from HIV-2. Early structural and biochemical studies showed that these residues contribute to substrate selectivity (27, 28), as well as inhibitor sensitivity (29), although most of these observations were made using investigational PI which were never licensed. More recently, crystallographic studies have linked three of the four residues (32, 47, and 82) to differential amprenavir (APV) and DRV sensitivities (30–33). However, the full extent of the four binding pocket differences has never been investigated using a live virus system, which encompasses all protease substrates in their native context.
In order to elucidate the contributions of HIV-2 protease amino acids Ile32, Val47, Met76, and Ile82 (Fig. 1B) to resistance to FDA-approved HIV-1 PI, we used replication-competent viruses in a single-cycle assay system. We constructed full-length molecular clones of HIV-2 containing all four substitutions (clone PRΔ4) as well as each substitution individually and compared the sensitivities of the resulting viruses to those of WT HIV-1 and HIV-2 for nine FDA-approved HIV-1 PI in culture. We also compared an energy-minimized model of the mutant HIV-2 protease structure to existing crystal structures of HIV proteases complexed with APV and DRV in order to determine a possible rationale for the differences in PI susceptibility. These studies provide insights into the determinants of PI sensitivity in HIV.
MATERIALS AND METHODS
Cell lines, antiretrovirals, and wild-type molecular clones.
293T/17 cells were purchased from the American Type Culture Collection (Manassas, VA), and MAGIC-5A indicator cells were kindly provided by Michael Emerman (Fred Hutchinson Cancer Research Center, Seattle, WA). 293T/17 and MAGIC-5A cells were maintained at 37°C with 5% CO2 in Dulbecco's modification of Eagle medium (DMEM; Mediatech, Manassas, VA) supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT), 4 mM l-glutamine, 50 U/ml of penicillin, and 50 μg/ml of streptomycin (Gibco Life Technologies Corp., Grand Island, NY).
The protease inhibitors amprenavir (APV), atazanavir (ATV), darunavir (DRV), indinavir (IDV), lopinavir (LPV), nelfinavir (NFV), ritonavir (RTV), saquinavir (SQV), and tipranavir (TPV) were obtained from the NIH AIDS Reagent Program. Master stocks (20 mM) were prepared in sterile high-performance liquid chromatography (HPLC)-grade dimethyl sulfoxide (DMSO; Alfa Aesar Co., Ward Hill, MA) and stored at −80°C. Serial dilutions of the drug were prepared in sterile distilled water and stored at −20°C.
Full-length infectious clones of HIV-1 (pNL4-3; group M, subtype B) and HIV-2 (pROD9; group A) were obtained from Bruce Chesebro (Rocky Mountain Laboratories, Hamilton, MT) and Michael Emerman, respectively.
Site-directed mutagenesis.
An HIV-2ROD9 molecular clone encoding protease substitutions corresponding to the amino acids found in wild-type HIV-1 (clone PRΔ4; I32V, V47I, M76L, and I82V) and clones containing each single substitution were engineered using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). All clones were sequenced across the entire genome to ensure that no extraneous mutations occurred during the mutagenesis process, and full-length plasmids were purified using a Hi-Speed Plasmid Maxi kit with endotoxin-free buffers (Qiagen Inc., Valencia, CA). Nucleotide sequences of the mutagenic primers are available upon request.
Virus production and titer assays.
Virus stocks were prepared by subjecting 293T/17 cells to chloroquine-mediated transfection of a standard quantity of plasmid DNA (14). Viral supernatants were harvested and clarified by 0.2-μm filtration. Single-cycle measurements of viral titers were performed in MAGIC-5A cells to ensure that mutant viruses were replication competent, using fresh, 2-fold serially diluted 293T/17 supernatants in complete DMEM supplemented with 20 μg/ml of DEAE-dextran (Sigma-Aldrich Co., St. Louis, MO). Culture monolayers were fixed in 1% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline (PBS) and then subjected to 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining as described previously (34). Focus-forming units (FFU) were counted by light microscopy. Mean titers (expressed as FFU/milliliter) were obtained from two independent replicates.
Protease inhibitor susceptibility assays.
The susceptibilities of each wild-type or mutant HIV-2 molecular clone were determined using a single-cycle assay as described previously (14). As described above, plasmid DNA was transfected into 293T/17 cells and then dosed with PI in half-log10 concentration increments. The resulting culture supernatants were plated onto MAGIC-5A indicator cells, and virus growth was quantified by β-galactosidase (β-Gal) cleavage of chlorophenol red-β-d-galactopyranoside (CPRG; BioShop Canada, Burlington, Ontario, Canada). All CPRG assay values were background subtracted to normalize for intrinsic β-Gal activity. Fifty percent effective concentrations (EC50s) were obtained from at least three sets of dose-response data using sigmoidal regression, log10 transformed, and tested for statistically significant differences by analysis of variance (ANOVA) with Sidak's test for multiple comparisons. All statistical analyses were carried out in Prism (version 6.0; GraphPad Software, Inc., San Diego, CA) using an α value of <0.05.
Structural studies.
Comparisons of existing crystal structures of HIV protease-inhibitor complexes were performed in UCSF Chimera (35). Structures used included HIV-1–APV (PDB code 3EKV [36]), HIV-2–APV (PDB code 3S45 [32]), HIV-1–DRV (PDB code 4DQB [37]), and HIV-2–DRV (PDB code 3EBZ [31]). To examine the structural changes responsible for drug sensitivity in the HIV-2 PRΔ4 mutant, amino acid changes I32V, V47I, M76L, and I82V were introduced into the wild-type HIV-2 protease-inhibitor complexes (PDB codes 3S45 and 3EBZ) using the Rotamers tool in Chimera. The resultant structures were energy minimized using YASARA (38). Similar structures were obtained using local energy minimizations performed in Chimera.
RESULTS
Generation of site-directed HIV-2 mutant viruses.
To determine the importance of the four amino acid differences in the active centers of HIV-1 and HIV-2 proteases, we used site-directed mutagenesis of the HIV-2ROD9 molecular clone to generate clone PRΔ4; this clone encodes protease substitutions I32V, V47I, M76L, and I82V, reflecting the amino acids found in WT HIV-1. We also generated HIV-2ROD9 clones encoding each substitution individually. When transfected into 293T/17 cells, all five mutant clones produced infectious virus. The mean titers of each mutant HIV-2 clone (4.4 to 4.6 log10 FFU per milliliter) were equivalent to that of WT HIV-2ROD9 (4.6 log10 FFU/ml).
Phenotypic PI susceptibilities of wild-type and mutant HIV-1 and HIV-2 strains.
Next, we compared the sensitivities of WT HIV-1NL4-3, WT HIV-2ROD9, and HIV-2 PRΔ4 (Fig. 2 and Table 1) to each of nine FDA-approved PI in a single-cycle assay. Relative to WT HIV-2ROD9, clone PRΔ4 was significantly more susceptible to all PI except SQV, to which HIV-1 and HIV-2 are equivalently sensitive. Changes in EC50s for PRΔ4 versus WT HIV-2 ranged between 2.6-fold and 60-fold, depending on the PI tested. The greatest increases in susceptibility between PRΔ4 and HIV-2 tended to occur for PI with which WT HIV-1 and HIV-2 EC50s are most different, including NFV (10-fold increase), DRV (26-fold), APV (>43-fold), and ATV (60-fold). Compared to those for HIV-1NL4-3, the EC50s for PRΔ4 were not statistically different, except for ATV and DRV, to which PRΔ4 was more susceptible (Fig. 2 and Table 1).
FIG 2.
PI sensitivities of wild-type HIV-1, wild-type HIV-2, and HIV-2 PRΔ4. Bars indicate mean EC50s from single-cycle assays in 293T/17 and MAGIC-5A indicator cells, comparing HIV-1NL4-3, HIV-2ROD9, and HIV-2ROD9 containing protease substitutions I32V, V47I, M76L, and I82V (PRΔ4). Error bars indicate standard deviations. Means and standard deviations were determined from three to eight independent assays. *, statistically significant (P < 0.05). ns, not significant. All differences between HIV-1NL4-3 and HIV-2ROD9 are statistically significant, except in the case of saquinavir.
TABLE 1.
Phenotypic PI sensitivities of wild-type HIV-1, wild-type HIV-2, and mutant HIV-2 strainsa
| Drug | PI sensitivity of: |
||||||
|---|---|---|---|---|---|---|---|
| HIV-1 |
HIV-2 |
||||||
| NL4-3 | ROD9 | I32V mutant | V47I mutant | M76L mutant | I82V mutant | PRΔ4 mutantb | |
| Amprenavir | 44 ± 13 | >1,000 | 550 ± 310 (1.8) | 470 ± 220 (2.1) | 660 ± 280 (1.5) | >1,000 (1.0) | 23 ± 17 (>43) |
| Atazanavir | 3.6 ± 3.0 | 66 ± 44 | 51 ± 18 (1.3) | 27 ± 3.4 (2.4) | 5.3 ± 2.1 (12) | 32 ± 5.9 (2.1) | 1.1 ± 1.1 (60) |
| Darunavir | 9.0 ± 3.9 | 58 ± 32 | 12 ± 0.7 (4.8) | 17 ± 3.3 (3.4) | 28 ± 7.8 (2.1) | 29 ± 2.4 (2.0) | 2.2 ± 1.3 (26) |
| Indinavir | 36 ± 23 | 150 ± 64 | 46 ± 21 (3.3) | 30 ± 25 (5.0) | 15 ± 6.8 (10) | 140 ± 140 (1.1) | 16 ± 9.8 (9.4) |
| Lopinavirc | 50 ± 29 | 123 ± 50 | 38 ± 22 (3.2) | 26 ± 8.7 (4.7) | 120 ± 64 (1.0) | 120 ± 98 (1.0) | 21 ± 10 (5.9) |
| Nelfinavir | 39 ± 18 | 490 ± 280 | 370 ± 90 (1.3) | 190 ± 86 (2.6) | 99 ± 69 (4.9) | 250 ± 130 (2.0) | 47 ± 4.9 (10) |
| Ritonavir | 61 ± 46 | 580 ± 240 | 290 ± 94 (1.6) | 510 ± 440 (1.3) | 240 ± 190 (2.4) | 620 ± 150 (1.0) | 160 ± 100 (3.6) |
| Saquinavir | 33 ± 33 | 31 ± 18 | 19 ± 12 (1.6) | 35 ± 7.7 (1.0) | 25 ± 12 (1.2) | 93 ± 71 (0.3) | 12 ± 6.1 (2.6) |
| Tipranavir | 250 ± 82 | >1,000 | >1,000 (1.0) | 260 ± 69 (>3.8) | 500 ± 270 (2.0) | >1,000 (1.0) | 120 ± 65 (>8.3) |
Data shown are means ± SD for EC50 (nanomolar concentration), with fold decrease compared to value wild-type strain HIV-2ROD9 in parentheses, for at least three independent dose-response assays. Bold indicates that the P value was <0.05 compared to the value for WT HIV-2ROD9 (by ANOVA of log10-transformed EC50; see Materials and Methods).
HIV-2 PRΔ4 contains the mutations I32V, V47I, M76L, and I82V.
We have previously reported lower lopinavir EC50s for WT HIV-1 and HIV-2 (14), using an earlier lot of the drug. In order to make valid comparisons, all lopinavir EC50s reported here are exclusively from assays using the same lot.
Relative to WT HIV-2, single amino acid substitutions I32V, V47I, M76L, and I82V conferred effects ranging from no change to a 12-fold decrease in EC50, depending on the PI tested (Table 1). None of these changes individually conferred significant increases in sensitivity to APV, RTV, or SQV. However, compared to WT HIV-2, one or more clones were significantly more susceptible to ATV, DRV, IDV, LPV, NFV, or TPV, in some cases with EC50s similar to those of WT HIV-1. However, no single change in HIV-2ROD9 protease conferred class-wide susceptibility equivalent to that of HIV-2 PRΔ4.
Structural comparison of HIV-1, HIV-2, and HIV-2 PRΔ4.
To help interpret the results of our culture-based assays, we examined the structures of HIV-1 and HIV-2 proteases cocrystallized with APV and DRV (31, 32, 35, 36). Importantly, HIV-2 is highly resistant to APV but susceptible to DRV; these compounds are closely related, the sole difference being the substitution of the tetrahydrofuran (THF) group of APV with bis-THF in DRV (Fig. 3).
FIG 3.
Chemical structures of amprenavir and darunavir. The locations of functional groups in the P1, P1′, P2, and P2′ sites of HIV-2 protease are shown for amprenavir; the same subsite orientation is used for the darunavir structure (adapted from reference 32).
In the APV structures, WT HIV-1 and HIV-2 show similar orientations of the inhibitor within the substrate-binding cleft (Fig. 4 and 5). Interactions between APV and amino acids in the P1 and P1′ sites of protease are conserved between HIV-1 and HIV-2, as are hydrogen-bonding interactions involving the catalytic aspartates D25 and D25′, and residues within the flap regions (and a coordinating water molecule) of the two enzyme-inhibitor complexes (Fig. 5). In contrast, subtle differences are apparent between HIV-1 and HIV-2 in the P2 and P2′ sites. These changes include loss of a potential C—H···π interaction between position 47 and the aniline ring of APV, increased distance between the main-chain carbonyl of G48 and the aniline group (from 3.5 to 3.8 Å for HIV-1 and HIV-2, respectively), lengthening of the distance between the main-chain carbonyl of D30′ and the THF ring (from 3.3 to 3.5 Å), and lengthening of the predicted hydrogen bond between the D30 main-chain carbonyl and the aniline amino group of APV (from 3.3 to 3.7 Å) (Fig. 4).
FIG 4.
Comparison of protein-inhibitor interactions for amprenavir (top) and darunavir (bottom) in complexes with wild-type HIV-1, wild-type HIV-2, and PRΔ4 HIV-2 proteases. Oxygen, nitrogen, and sulfur atoms are depicted in red, blue, and orange, respectively. The HIV-2 PRΔ4 structures were made by introducing amino acid changes I32V, V47I, M76L, and I82V into the wild-type HIV-2 protease-inhibitor complexes (PDB codes 3S45 and 3EBZ) using the Rotamers tool in Chimera and energy minimized using YASARA. Water molecules coordinated by amprenavir or darunavir and the main-chain nitrogen atoms of I50 and I50′ are shown as red spheres. The energy-minimized model of PRΔ4 HIV-2 with darunavir (lower right) did not contain a water molecule at this position. Dotted red lines indicate hydrogen bonds. Dotted black lines indicate potential hydrophobic and C—H···O interactions. Atomic distances are shown in angstroms.
FIG 5.
Positioning of amprenavir and amino acids proximal to the P1 and P1′ sites of wild-type HIV-1, wild-type HIV-2, and PRΔ4 HIV-2 proteases. Bottom-row views are rotated 180° along the y axis relative to top-row views for all structures. Amprenavir is shown in purple. Residues 76 to 84 and 76′ to 84′ are shown as tan spheres. Yellow sticks indicate residues 32:32′ and 47 to 50:47′ to 50′. Cyan sticks indicate residues 27 to 30:27′ to 30′.
We then examined the structural changes responsible for APV sensitivity in HIV-2 PRΔ4 using an energy-minimized model of the HIV-2 structure containing the PRΔ4 substitutions. Compared to WT HIV-2, residues in the P1 and P1′ pockets of the HIV-2 PRΔ4 mutant maintain similar interactions with the benzyl and isopropyl groups of APV, respectively (Fig. 5), whereas P2 and P2′ show several alterations that could potentially influence APV binding. In the PRΔ4 model, the side chain of V47′, the main-chain carbonyl oxygen of D30′, and the main-chain nitrogen atoms of D29′ and D30′ are in closer proximity to the THF moiety of APV (differences of 0.6, 0.1, 0.2, and 0.1 Å, respectively, relative to WT HIV-2) (Fig. 4). In the P2′ pocket, substitution of V47 in WT HIV-2 with the longer isoleucine residue of the PRΔ4 mutant brings the side chain of residue 47 0.8 Å closer to the aniline ring, potentially forming a new C—H···π interaction between the PRΔ4 mutant protease and APV. In addition, in the PRΔ4 model, the main-chain carbonyl group of D30 is rotated approximately 40° relative to the WT HIV-2 structure, resulting in a conformation similar to the one observed in WT HIV-1 protease (Fig. 4). This rearrangement results in a shorter hydrogen bonding distance between the carbonyl oxygen and the aniline NH2 of APV (from 3.7 to 3.2 Å) (Fig. 4).
In contrast to APV, complexes of HIV-1 and HIV-2 protease with DRV show little difference between the two HIV types. Protein-ligand interactions believed to be important for DRV activity (31), including hydrogen bonds and C—H···π and C—H···O interactions, are well conserved between HIV-1 and HIV-2, with differences of ≤0.2 Å between the two structures (Fig. 4). PRΔ4 protease interacts with DRV in a manner similar to both WT HIV-1 and WT HIV-2 enzymes. These findings are consistent with data from our group and others showing that DRV exhibits substantial activity against WT HIV-1, WT HIV-2, and HIV-2 PRΔ4 in cell culture (Fig. 2).
DISCUSSION
We and others have previously shown that wild-type HIV-2 is at least partially resistant, in vitro, to the majority of PI FDA approved for the treatment of HIV-1, retaining clinically useful susceptibility only to SQV, LPV, and DRV (13–20). Although it has long been known that the active centers of HIV-1 and HIV-2 proteases differ by only four amino acids (23), this observation has never been fully investigated. We report, for the first time, that replacing four active-site amino acid residues in HIV-2 protease with the corresponding amino acids from HIV-1 (I32V, V47I, M76L, and I82V) results in a replication-competent virus which exhibits a pattern of class-wide PI sensitivity comparable to that of HIV-1.
A few studies have suggested that the four differing active-center residues may be responsible for the differences in PI susceptibility between HIV-1 and HIV-2. Modeling studies by Gustchina et al. revealed that differences in the affinity of HIV-1 and HIV-2 proteases to two aspartic PI appeared to be due in part to I32V (27). Sardana and colleagues mutated the four amino acid positions in HIV-1 protease to the corresponding HIV-2 amino acids and performed enzyme activity and inhibition studies using purified proteases in the presence of three inhibitors: L-689,502, L-731,723, and RO 31-8959 (later licensed as SQV) (29). Although their results supported the findings of Gustchina et al., the authors noted that substitutions at residues 47, 76, and 82 had relatively minor effects on protease inhibition by these compounds. In addition, the HIV-1 mutant protease containing all four changes was only slightly resistant to the inhibitors, leading to the conclusion that the four active-site residues would be insufficient to explain the differences in inhibitor sensitivity between HIV-1 and HIV-2 proteases. This observation was further explored by Hoog et al., who solved the structure of an HIV-1 protease containing V32I, I47V, and V82I, complexed with the tripeptide analogue inhibitor SB203386 and compared it to previously solved structures of WT HIV-1 and simian immunodeficiency virus (SIV) complexed with the same inhibitor (28). They concluded that ligand specificity was imparted not only by the three active-site residues tested but also by changes outside the active center.
More recent studies have explored the structural elements involved in differential APV and DRV susceptibilities (30–33). Tie and colleagues examined structural and biochemical features of proteases from HIV-1, HIV-2, and HIV-1 containing V32I, I47V, and V82I (32). Although the overall structure of the HIV-1 mutant protease more closely resembled that of WT HIV-1 protease, its biochemical parameters were similar to those of the HIV-2 protease, and the interactions observed in their enzyme-APV structure for the HIV-1 mutant were also more similar to those seen in the HIV-2 structure. In keeping with the observation that HIV-1 and HIV-2 display relatively similar sensitivities to DRV, this study noted that hydrogen bond interactions in the DRV-containing structures were very similar between all three proteases. This and other studies have identified a number of electrostatic and hydrophobic interactions thought to be important in the observed differences in sensitivity between SQV, DRV, and APV (30–33). Together, these studies reveal that small rearrangements resulting from one, two, or three amino acid substitutions in the substrate-binding pockets of HIV proteases result in changes in the internal interactions, which may lead to differing patterns of PI sensitivity.
No previous study has examined the drug sensitivity of HIV-2 variants containing these four amino acid changes, either alone or in combination, using a live-virus assay system. Our results demonstrate that in HIV-2 protease, amino acid changes I32V, V47I, M76L, and I82V cooperate to confer a level of PI susceptibility greater than the sum of each individual change. HIV-2 clone PRΔ4, containing the corresponding HIV-1 amino acids at positions 32, 47, 76, and 82, exhibits PI susceptibility either equivalent to, or better than, that of HIV-1. The mechanisms causing the HIV-2 PRΔ4 clone to exhibit lower EC50s with ATV and DRV than those seen in HIV-1NL4-3 are unclear and warrant further study. In HIV-1, PI resistance-associated substitutions occur throughout the protease enzyme and can restore viral fitness and/or enhance the effects of primary resistance mutations. Amino acids outside the HIV-2 protease active site may cause subtle changes in conformation compared to HIV-1, which might impact PI binding affinity. Future studies of different HIV-2 isolates from groups A and B with I32V, V47I, M76L, and I82V substitutions but with differing background polymorphisms in protease could help address these possibilities.
There are a number of other potential limitations to our study. We did not compare in vitro enzymatic activity and PI susceptibility of the purified HIV-2 PRΔ4 enzyme to those of WT HIV-2 protease; however, we contend that cell culture-based assays are a more realistic approximation of in vivo PI susceptibility. We also did not test combinations of two or three amino acid changes within the four active-site residues, so we cannot exclude the possibility that other combinations of mutations at these or other sites would similarly “resensitize” HIV-2 to these compounds. In addition, our site-directed mutations were made in only one strain, HIV-2ROD9, but the high level of amino acid conservation at these sites makes it less likely that other strains would show dramatically different results. We did not generate and test the “reverse” PRΔ4 mutant in HIV-1 (that is, HIV-1 containing the corresponding HIV-2 amino acids). However, many previous studies have shown that single amino acid changes at Val32, Ile47, Leu76, and Val82 confer PI resistance (summarized in references 26, 39, and 40). According to the Stanford HIV Drug Resistance Database (http://hivdb.stanford.edu), V32I is a major PI resistance-associated substitution, conferring resistance to fosamprenavir (FPV; a prodrug of APV), ATV, DRV, IDV, LPV, and NFV. Similarly, I47V is a major resistance-associated mutation conferring decreased susceptibility to FPV, DRV, IDV, LPV, NFV, and TPV, respectively. Although neither L76M nor V82I is known to cause PI resistance in HIV-1, other substitutions at these positions are associated with intermediate to high-level resistance to multiple PI. The combined reverse PRΔ4 mutant in HIV-1 (V32I-I47V-L76M-V82I) is predicted by the Stanford HIV Drug Resistance Database to have high-level resistance (>11-fold) to FPV, intermediate resistance (3-10-fold) to DRV, IDV, LPV, NFV, and TPV, and low-level resistance (<3-fold) to ATV and to retain SQV susceptibility.
In agreement with previously published studies using an earlier structure of HIV-1 protease-APV (PDB code 3NU3) (30–32), our structural analyses suggest that HIV-2 resistance to APV results from a diminution or loss of stabilizing interactions between protease and the THF and aniline groups of the inhibitor. It appears that the PRΔ4 mutations help to stabilize APV in the P2 and P2′ pockets of HIV-2 protease, possibly accounting for the increased susceptibility of HIV-2 PRΔ4 to APV in culture. The differences in DRV sensitivity between HIV-1 and HIV-2 are relatively small in comparison to the differences in their sensitivities to APV, lending support to the observation that HIV-1, HIV-2, and HIV-2 PRΔ4 proteases interact similarly with DRV (31–33). Although crystallographic studies can provide important clues into the structural mechanisms behind differential PI susceptibilities, there are important limitations to this approach. First, due to the dearth of X-ray crystallographic structures of HIV-2 complexed with PI, a comprehensive analysis of the structural basis for the differences in PI susceptibility between HIV-1 and HIV-2 is not possible. In addition, our comparison uses models of PRΔ4 rather than a solved structure of the mutant enzyme and thus contains inherent uncertainty. In particular, there is a coordinating water missing from the flap region of the active site in the PRΔ4-DRV model, which could potentially reveal a problem with the model. However, we believe this to be an artifact of the energy minimization process, as the relevant main-chain nitrogen atoms from the PRΔ4 protease, and the oxygen atoms of DRV that should coordinate this water molecule, are appropriately positioned to coordinate a water molecule at this site. Finally, although crystal structures demonstrate structural changes in the enzyme-inhibitor interactions caused by amino acid substitutions, they may not adequately account for in changes in enzyme dynamic and thermodynamic properties that result from these mutations.
To date, no antiretroviral compounds have been developed or licensed for the treatment of HIV-2; as a result, HIV-2 treatment relies solely on drugs developed for HIV-1. Although the course of HIV-2 infection is relatively protracted compared to that of HIV-1, with lower viral loads and slower decline in CD4+ T cells (41, 42), without the benefit of antiretroviral therapy (ART), a substantial proportion of infected individuals will progress to AIDS (43). Because of this, HIV-2 is a significant public health challenge in West Africa, where the virus is endemic (44). Although PI-based regimens are the mainstay of HIV-2 treatment in resource-limited settings, only a third of FDA-approved PI are HIV-2 active at clinically useful concentrations, and only one of those agents (LPV/RTV; Kaletra or Aluvia) is widely available for clinical use in resource-limited settings.
Our findings show that four residues in the protease binding pocket are the primary determinants of intrinsic PI resistance in HIV-2, enhancing our overall understanding of the genetic basis of PI susceptibility. Further, these results provide evidence that subtle structural changes imparted by a limited number of residues can cause dramatic functional differences between homologous enzymes from two closely related viruses.
ACKNOWLEDGMENTS
The University of Washington-Dakar HIV-2 Study Group includes Moussa Seydi, Papa Salif Sow, Selly Ba, Macoumba Toure, Fatima Sall, Fatou Traore, Khadim Faye, Ousseynou Ndiaye, Marie Pierre Sy, Bintou Diaw, Mbaye Ndoye, Amadou Bale Diop, and Marianne Fadam Diome, all from the Service des Maladies Infectieuses Ibrahima Diop Mar, Centre Hospitalier Universitaire de Fann, Universite Cheikh Anta Diop de Dakar, Dakar, Senegal; Alassane Niang, ElHadji Ibrahima Sall, Ousseynou Cisse, Ibrahima Tito Tamba, Jean Phillippe Diatta, Raphael Bakhoum, Jacque Francois Sambou, and Juliette Gomis, all from the Region Medicale de Ziguinchor, Ziguinchor, Casamance, Senegal; and Stephen Hawes, Noelle Benzekri, Robert Coombs, Ming Chang, John Lin, Nancy Kiviat, James Mullins, Sally Leong, and Vincent Wu, all from the University of Washington.
Funding Statement
The University of Washington Center for AIDS Research (CFAR) is an NIH-funded program.
REFERENCES
- 1.Kohl NE, Emini EA, Schleif WA, Davis LJ, Heimbach JC, Dixon RA, Scolnick EM, Sigal IS. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci U S A 85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debouck C, Gorniak JG, Strickler JE, Meek TD, Metcalf BW, Rosenberg M. 1987. Human immunodeficiency virus protease expressed in Escherichia coli exhibits autoprocessing and specific maturation of the gag precursor. Proc Natl Acad Sci U S A 84:8903–8906. doi: 10.1073/pnas.84.24.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wlodawer A, Miller M, Jaskolski M, Sathyanarayana BK, Baldwin E, Weber IT, Selk LM, Clawson L, Schneider J, Kent SB. 1989. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science 245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 4.Meek TD, Lambert DM, Dreyer GB, Carr TJ, Tomaszek TA Jr, Moore ML, Strickler JE, Debouck C, Hyland LJ, Matthews TJ, Metcalf BW, Petteway SR. 1990. Inhibition of HIV-1 protease in infected T-lymphocytes by synthetic peptide analogues. Nature 343:90–92. doi: 10.1038/343090a0. [DOI] [PubMed] [Google Scholar]
- 5.Ingr M, Uhlikova T, Strisovsky K, Majerova E, Konvalinka J. 2003. Kinetics of the dimerization of retroviral proteases: the “fireman's grip” and dimerization. Protein Sci 12:2173–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gustchina A, Weber IT. 1991. Comparative analysis of the sequences and structures of HIV-1 and HIV-2 proteases. Proteins 10:325–339. doi: 10.1002/prot.340100406. [DOI] [PubMed] [Google Scholar]
- 7.Louis JM, Ishima R, Torchia DA, Weber IT. 2007. HIV-1 protease: structure, dynamics, and inhibition. Adv Pharmacol 55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 8.Mulichak AM, Hui JO, Tomasselli AG, Heinrikson RL, Curry KA, Tomich CS, Thaisrivongs S, Sawyer TK, Watenpaugh KD. 1993. The crystallographic structure of the protease from human immunodeficiency virus type 2 with two synthetic peptidic transition state analog inhibitors. J Biol Chem 268:13103–13109. [PubMed] [Google Scholar]
- 9.Tong L, Pav S, Pargellis C, Do F, Lamarre D, Anderson PC. 1993. Crystal structure of human immunodeficiency virus (HIV) type 2 protease in complex with a reduced amide inhibitor and comparison with HIV-1 protease structures. Proc Natl Acad Sci U S A 90:8387–8391. doi: 10.1073/pnas.90.18.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Li Y, Chen E, Hall DL, Darke PL, Culberson C, Shafer JA, Kuo LC. 1994. Crystal structure at 1.9-A resolution of human immunodeficiency virus (HIV) II protease complexed with L-735,524, an orally bioavailable inhibitor of the HIV proteases. J Biol Chem 269:26344–26348. [PubMed] [Google Scholar]
- 11.Menéndez-Arias L, Tozser J. 2008. HIV-1 protease inhibitors: effects on HIV-2 replication and resistance. Trends Pharmacol Sci 29:42–49. doi: 10.1016/j.tips.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Patterson CE, Seetharam R, Kettner CA, Cheng YS. 1992. Human immunodeficiency virus type 1 and type 2 protease monomers are functionally interchangeable in the dimeric enzymes. J Virol 66:1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brower ET, Bacha UM, Kawasaki Y, Freire E. 2008. Inhibition of HIV-2 protease by HIV-1 protease inhibitors in clinical use. Chem Biol Drug Des 71:298–305. doi: 10.1111/j.1747-0285.2008.00647.x. [DOI] [PubMed] [Google Scholar]
- 14.Raugi DN, Smith RA, Ba S, Toure M, Traore F, Sall F, Pan C, Blankenship L, Montano A, Olson J, Dia Badiane NM, Mullins JI, Kiviat NB, Hawes SE, Sow PS, Gottlieb GS. 2013. Complex patterns of protease inhibitor resistance among antiretroviral treatment-experienced HIV-2 patients from Senegal: implications for second-line therapy. Antimicrob Agents Chemother 57:2751–2760. doi: 10.1128/AAC.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desbois D, Roquebert B, Peytavin G, Damond F, Collin G, Benard A, Campa P, Matheron S, Chene G, Brun-Vezinet F, Descamps D. 2008. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrob Agents Chemother 52:1545–1548. doi: 10.1128/AAC.01284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. 2004. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir Ther 9:57–65. [PubMed] [Google Scholar]
- 17.Masse S, Lu X, Dekhtyar T, Lu L, Koev G, Gao F, Mo H, Kempf D, Bernstein B, Hanna GJ, Molla A. 2007. In vitro selection and characterization of human immunodeficiency virus type 2 with decreased susceptibility to lopinavir. Antimicrob Agents Chemother 51:3075–3080. doi: 10.1128/AAC.00146-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavaco-Silva J, Aleixo MJ, Van Laethem K, Faria D, Valadas E, Goncalves MD, Gomes P, Vandamme AM, Cunha C, Camacho RJ. 13 September 2012. Mutations selected in HIV-2-infected patients failing a regimen including atazanavir. J Antimicrob Chemother doi: 10.1093/jac/dks363. [DOI] [PubMed] [Google Scholar]
- 19.Bénard A, Damond F, Campa P, Peytavin G, Descamps D, Lascoux-Combes C, Taieb A, Simon F, Autran B, Brun-Vezinet F, Chene G, Matheron S. 2009. Good response to lopinavir/ritonavir-containing antiretroviral regimens in antiretroviral-naive HIV-2-infected patients. AIDS 23:1171–1173. doi: 10.1097/QAD.0b013e32832949f0. [DOI] [PubMed] [Google Scholar]
- 20.Rodés B, Sheldon J, Toro C, Jimenez V, Alvarez MA, Soriano V. 2006. Susceptibility to protease inhibitors in HIV-2 primary isolates from patients failing antiretroviral therapy. J Antimicrob Chemother 57:709–713. doi: 10.1093/jac/dkl034. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Pescador R, Power MD, Barr PJ, Steimer KS, Stempien MM, Brown-Shimer SL, Gee WW, Renard A, Randolph A, Levy JA, Dina D, Luciw PA. 1985. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2). Science 227:484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- 22.Guyader M, Emerman M, Sonigo P, Clavel F, Montagnier L, Alizon M. 1987. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature 326:662–669. doi: 10.1038/326662a0. [DOI] [PubMed] [Google Scholar]
- 23.Tözsér J, Blaha I, Copeland TD, Wondrak EM, Oroszlan S. 1991. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett 281:77–80. doi: 10.1016/0014-5793(91)80362-7. [DOI] [PubMed] [Google Scholar]
- 24.Tözsér J, Weber IT, Gustchina A, Blaha I, Copeland TD, Louis JM, Oroszlan S. 1992. Kinetic and modeling studies of S3-S3′ subsites of HIV proteinases. Biochemistry 31:4793–4800. doi: 10.1021/bi00135a008. [DOI] [PubMed] [Google Scholar]
- 25.Tomasselli AG, Hui JO, Sawyer TK, Staples DJ, Bannow C, Reardon IM, Howe WJ, DeCamp DL, Craik CS, Heinrikson RL. 1990. Specificity and inhibition of proteases from human immunodeficiency viruses 1 and 2. J Biol Chem 265:14675–14683. [PubMed] [Google Scholar]
- 26.Wensing AM, Calvez V, Gunthard HF, Johnson VA, Paredes R, Pillay D, Shafer RW, Richman DD. 2014. 2014 update of the drug resistance mutations in HIV-1. Top Antivir Med 22:642–650. [PMC free article] [PubMed] [Google Scholar]
- 27.Gustchina A, Weber IT, Wlodawer A. 1991. Molecular modeling of the HIV-2 protease. Adv Exp Med Biol 306:549–553. doi: 10.1007/978-1-4684-6012-4_75. [DOI] [PubMed] [Google Scholar]
- 28.Hoog SS, Towler EM, Zhao B, Doyle ML, Debouck C, Abdel-Meguid SS. 1996. Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity. Biochemistry 35:10279–10286. doi: 10.1021/bi960179j. [DOI] [PubMed] [Google Scholar]
- 29.Sardana VV, Schlabach AJ, Graham P, Bush BL, Condra JH, Culberson JC, Gotlib L, Graham DJ, Kohl NE, LaFemina RL, Schneider CL, Wolanski BS, Wolfgang JA, Emini EA. 1994. Human immunodeficiency virus type 1 protease inhibitors: evaluation of resistance engendered by amino acid substitutions in the enzyme's substrate binding site. Biochemistry 33:2004–2010. doi: 10.1021/bi00174a005. [DOI] [PubMed] [Google Scholar]
- 30.Shen CH, Wang YF, Kovalevsky AY, Harrison RW, Weber IT. 2010. Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters. FEBS J 277:3699–3714. doi: 10.1111/j.1742-4658.2010.07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovalevsky AY, Louis JM, Aniana A, Ghosh AK, Weber IT. 2008. Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease. J Mol Biol 384:178–192. doi: 10.1016/j.jmb.2008.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tie Y, Wang YF, Boross PI, Chiu TY, Ghosh AK, Tozser J, Louis JM, Harrison RW, Weber IT. 2012. Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors. Protein Sci 21:339–350. doi: 10.1002/pro.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kar P, Knecht V. 2012. Origin of decrease in potency of darunavir and two related antiviral inhibitors against HIV-2 compared to HIV-1 protease. J Phys Chem B 116:2605–2614. doi: 10.1021/jp211768n. [DOI] [PubMed] [Google Scholar]
- 34.Smith RA, Anderson DJ, Pyrak CL, Preston BD, Gottlieb GS. 2009. Antiretroviral drug resistance in HIV-2: three amino acid changes are sufficient for classwide nucleoside analogue resistance. J Infect Dis 199:1323–1326. doi: 10.1086/597802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.King NM, Prabu-Jeyabalan M, Bandaranayake RM, Nalam MN, Nalivaika EA, Ozen A, Haliloglu T, Yilmaz NK, Schiffer CA. 2012. Extreme entropy-enthalpy compensation in a drug-resistant variant of HIV-1 protease. ACS Chem Biol 7:1536–1546. doi: 10.1021/cb300191k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mittal S, Cai Y, Nalam MN, Bolon DN, Schiffer CA. 2012. Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease. J Am Chem Soc 134:4163–4168. doi: 10.1021/ja2095766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieger E, Vriend G. 2014. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. 2010. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob Agents Chemother 54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menéndez-Arias L. 2013. Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments. Antiviral Res 98:93–120. doi: 10.1016/j.antiviral.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Simon F, Matheron S, Tamalet C, Loussert-Ajaka I, Bartczak S, Pepin JM, Dhiver C, Gamba E, Elbim C, Gastaut JA, Saimot AG, Brun-Vezinet F. 1993. Cellular and plasma viral load in patients infected with HIV-2. AIDS 7:1411–1417. doi: 10.1097/00002030-199311000-00002. [DOI] [PubMed] [Google Scholar]
- 42.Whittle H, Morris J, Todd J, Corrah T, Sabally S, Bangali J, Ngom PT, Rolfe M, Wilkins A. 1994. HIV-2-infected patients survive longer than HIV-1-infected patients. AIDS 8:1617–1620. doi: 10.1097/00002030-199411000-00015. [DOI] [PubMed] [Google Scholar]
- 43.Matheron S, Pueyo S, Damond F, Simon F, Lepretre A, Campa P, Salamon R, Chene G, Brun-Vezinet F. 2003. Factors associated with clinical progression in HIV-2 infected-patients: the French ANRS cohort. AIDS 17:2593–2601. doi: 10.1097/00002030-200312050-00006. [DOI] [PubMed] [Google Scholar]
- 44.Ariën KK, Abraha A, Quinones-Mateu ME, Kestens L, Vanham G, Arts EJ. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J Virol 79:8979–8990. doi: 10.1128/JVI.79.14.8979-8990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]