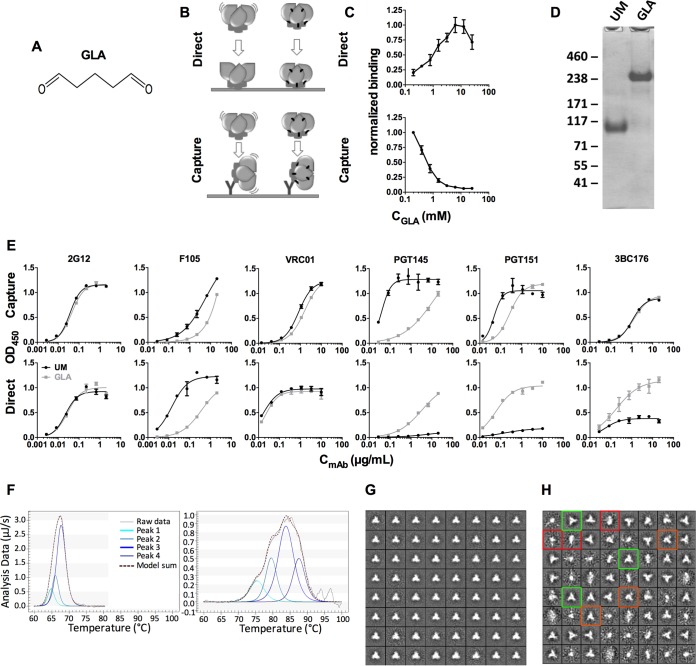
FIG 1.
Direct coating ELISA and establishment of optimal cross-linking conditions. (A) Structure of the GLA cross-linker. (B) Depiction of the ELISA formats:  indicates conformational instability, and black dashes indicate GLA cross-links within the trimer. (C) ELISA data corresponding to panel B; PGT145 binding to BG505 trimers cross-linked by different GLA concentrations and assayed by direct binding or capture ELISA. (D) Reducing SDS-PAGE analysis of BG505 trimers, either unmodified (UM) or cross-linked with 7.5 mM GLA for 5 min. (E) Unmodified and GLA cross-linked trimers were analyzed for antibody binding by direct coating and capture ELISA. Color development times differ for each antibody and between the ELISA formats to yield comparable endpoint signals, and hence the plots are not representations of the relative exposure of different epitopes on the trimers. Each datum point represents the mean of triplicates ± standard error of the mean (SEM). Data are representative of at least two independent experiments. (F) Unmodified and GLA cross-linked material was analyzed by DSC. Modeled peaks from a non-two-state fit as well as their sum are indicated. Data from the DSC analysis are listed, including individual peaks of a non-two-state model. (G and H) Negative-stain EM analysis of unmodified and GLA-treated BG505 trimers. 2D class averages are shown for unmodified (G) and GLA cross-linked (H) trimers. Exemplary closed native-like trimers (green squares), open native-like trimers (orange squares), and nonnative-like trimers (red squares) are marked in panel H. In total, >95% of trimers in panel G were correctly folded, whereas 2,466 of 4,561 (54%) of the GLA cross-linked trimers in panel H were correctly folded.
indicates conformational instability, and black dashes indicate GLA cross-links within the trimer. (C) ELISA data corresponding to panel B; PGT145 binding to BG505 trimers cross-linked by different GLA concentrations and assayed by direct binding or capture ELISA. (D) Reducing SDS-PAGE analysis of BG505 trimers, either unmodified (UM) or cross-linked with 7.5 mM GLA for 5 min. (E) Unmodified and GLA cross-linked trimers were analyzed for antibody binding by direct coating and capture ELISA. Color development times differ for each antibody and between the ELISA formats to yield comparable endpoint signals, and hence the plots are not representations of the relative exposure of different epitopes on the trimers. Each datum point represents the mean of triplicates ± standard error of the mean (SEM). Data are representative of at least two independent experiments. (F) Unmodified and GLA cross-linked material was analyzed by DSC. Modeled peaks from a non-two-state fit as well as their sum are indicated. Data from the DSC analysis are listed, including individual peaks of a non-two-state model. (G and H) Negative-stain EM analysis of unmodified and GLA-treated BG505 trimers. 2D class averages are shown for unmodified (G) and GLA cross-linked (H) trimers. Exemplary closed native-like trimers (green squares), open native-like trimers (orange squares), and nonnative-like trimers (red squares) are marked in panel H. In total, >95% of trimers in panel G were correctly folded, whereas 2,466 of 4,561 (54%) of the GLA cross-linked trimers in panel H were correctly folded.