Figure 1.
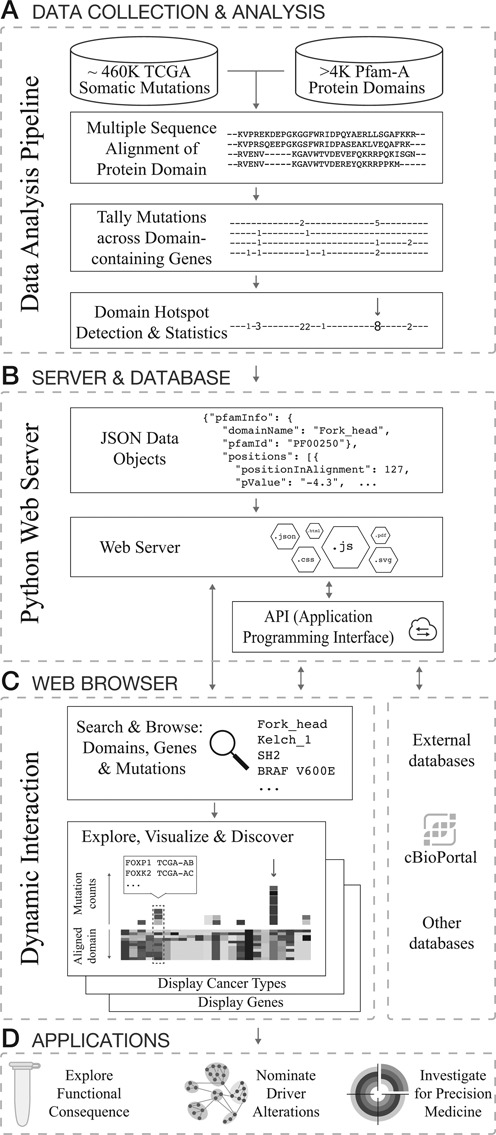
Database construction and software architecture of mutationaligner.org. (A) Integrated analysis of cancer mutations from TCGA and protein domains from Pfam-A. (B) Identified domain mutation hotspots and analysis results are stored and served from a python webserver via a documented Application Programming Interface (API, see main text). The webserver also provides the files required for the single page javascript application. (C) Analysis results are provided interactively in the user's web browser (client-side) through a rich, single page javascript application. (D) Examples of potential follow-on applications based on observations made using MutationAligner.
