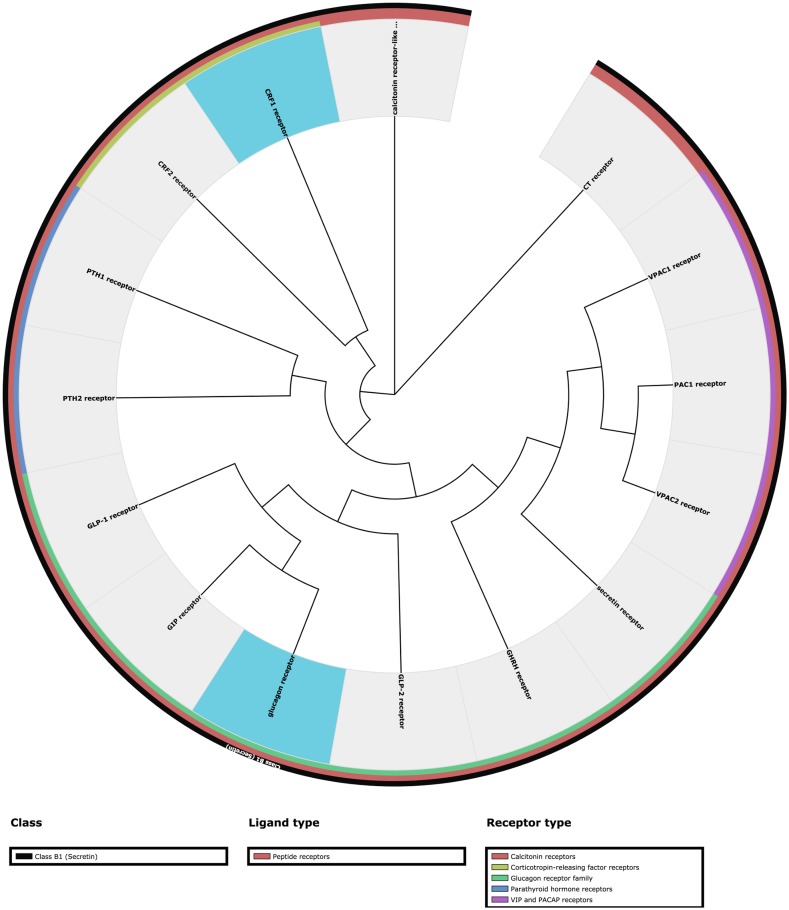
Figure 3.
This figure shows the human Class B1 (Secretin family) GPCRs. Custom Phylogenetic trees can swiftly be calculated under the Sequences tools menu. The interpretation of the trees is guided by coloured bars next to the receptor names that list the GPCR class, ligand type and receptor family, respectively. Receptors that have been crystallized can be highlighted with a blue background, and other background colours can be manually assigned to produce custom illustrations. Pre-generated phylogenetic trees available in the Structure statistics page show the structural coverage within each GPCR Class. For Class A, the consensus sequences are used for receptor families without a structure, whereas all receptor subtypes are included for the other Classes.