Abstract
Post-marketing drug withdrawals can be associated with various events, ranging from safety issues such as reported deaths or severe side-effects, to a multitude of non-safety problems including lack of efficacy, manufacturing, regulatory or business issues. During the last century, the majority of drugs voluntarily withdrawn from the market or prohibited by regulatory agencies was reported to be related to adverse drug reactions. Understanding the underlying mechanisms of toxicity is of utmost importance for current and future drug discovery. Here, we present WITHDRAWN, a resource for withdrawn and discontinued drugs publicly accessible at http://cheminfo.charite.de/withdrawn. Today, the database comprises 578 withdrawn or discontinued drugs, their structures, important physico-chemical properties, protein targets and relevant signaling pathways. A special focus of the database lies on the drugs withdrawn due to adverse reactions and toxic effects. For approximately one half of the drugs in the database, safety issues were identified as the main reason for withdrawal. Withdrawal reasons were extracted from the literature and manually classified into toxicity types representing adverse effects on different organs. A special feature of the database is the presence of multiple search options which will allow systematic analyses of withdrawn drugs and their mechanisms of toxicity.
INTRODUCTION
Efficacy and safety are two decisive factors that affect the viability of a chemical entity while furthering in the drug discovery pipeline. Consequently, the financial burden on pharmaceutical companies grows higher when the chemical entities tend to fail in late stages of clinical trials (1). However, a significant number of new chemical entities (NCEs) were recalled from the market post to their regulatory approval due to various reasons ranging from inefficiency to severe side-effects to financial and regulatory concerns. Adverse drug reactions (ADRs) not only account for market withdrawals but also for changes in labels or introduction of new black-box warnings for prescription drugs (2). ADRs can be interpreted either as primary effects elicited after modulation of the therapeutic (or primary) target or unintended effects due to interactions with off-targets. In few instances, the primary target is expressed in multiple organs and simultaneously targeted, leading to the therapeutic effect in the target tissue and unwanted effects in other tissues.
A well-known class of drugs that cause adverse reactions due to their activity at primary target are antiarrhythmic drugs, the benefits of which are, in few cases, hindered due to aggravation of arrhythmia which is the indication being treated (3). This effect is due to modulation of the alpha subunit of a potassium ion channel (human Ether-à-go-go-related gene, hERG), which is primarily associated with regulation of cardiac action potentials (4). The hERG channel is also a prominent off-target example whose unintended modulation can cause severe side-effects. This has ultimately lead to market withdrawal of drugs inhibiting the hERG channel, a classical example being the withdrawal of the antihistaminic drug terfenadine due to severe arrhythmias and death (5).
Although there is much progress in elucidation and understanding of the mechanisms leading to drug related toxic effects, gaining clearer insights about these effects at cellular and biochemical level is much needed to appropriately adjust or reinvent the development strategies so as to overcome the attrition during clinical trial phases of drug discovery and withdrawal after drug approval (6–8). This toxicological knowledge could be used to develop a panel of relevant in vitro assays that could mechanistically examine the effects and profile the propensity of drugs to cause ADRs (9). In contrast to the majority of ADR cases which are relatively frequent and mostly dose-dependent, few side-effects are idiosyncratic drug reactions (IADRs), i.e. the extremely rare drug reactions which occur unpredictably in a population. The target organs that are most commonly associated with idiosyncratic events include liver, cardiovascular and central nervous systems (10–12). Hepatocellular and cholestatic drug-induced liver injury (DILI), liver failure and hepatic necrosis are the common patterns of IADRs associated with the liver. Limited knowledge exists to understand the underlying mechanisms of such IADRs. However, it is apparent that IADRs develop via complex mechanisms which are subjective to both differential patient responses and drug combination effects that result from simultaneous triggering of multiple off-targets (13). Factors associated with differential patient responses include genetic attributes like single nucleotide polymorphisms (SNPs) and mutations, and non-genetic attributes such as gender, age and co-treatments (14). Drug-induced events are a result of various effects ranging from direct activity on organs (e.g. on cardiovascular systems) to reactivity of active metabolites of drugs to interactions with biological transporters (15).
Over the decades, drug regulatory agencies, pharmaceutical companies and various clinical studies have reported the events of drug withdrawals due to side-effects (16–18). About 2.3 million adverse event reports were collected against ∼6000 marketed drugs between 1969 and 2002 (19). Yet, only a small proportion (75 drugs; ∼1%) of these marketed drugs were withdrawn during this period. Another study reported that ∼95 drugs were documented to be withdrawn due to death as the primary reason between 1950 and 2013 (17). However, not all of these drugs were withdrawn world-wide. Most drugs were reported to be withdrawn in the Unites States and European countries.
Several public resources contain information relevant to drug withdrawals (e.g. websites from regulatory agencies, World Health Organization's consolidated list for withdrawn drugs and scientific literature). However, in many cases, the information is hidden in regulatory documents and not easily accessible, impeding comprehensive analyses. Furthermore, there exists no single resource reporting a complete list of drugs withdrawn due to safety concerns. In order to allow access to a variety of information related to drug withdrawals as well as shed light on the mechanisms of ADRs, we here present WITHDRAWN—a resource for withdrawn and discontinued drugs. We collected a list of more than 500 drugs/drug products, which were withdrawn or discontinued in at least one country, and assembled information regarding their molecular targets, pathways and toxicities. For approximately half of the drugs, extensive literature search revealed that toxic events are associated with the withdrawal. Thus, WITHDRAWN can be seen as a platform to understand the mechanisms for severe ADRs due to primary and off-target interactions of drugs, simultaneous perturbation of complex biological pathways and genetic polymorphisms (SNPs). Furthermore, it provides multiple search options to systematically analyse molecules of interest by performing different types of molecular similarity search across the database's drugs and can be a valuable resource for scientists in the drug development and toxicity prediction field.
MATERIALS AND METHODS
Withdrawn and discontinued drugs
A number of resources including the drug collections from the U.S. Food and Drug Administration (FDA; http://www.fda.gov/), the European Medicines Agency (EMA; http://www.ema.europa.eu/ema/), peer-reviewed literature (17), public databases such as DrugBank (20), e-Drug3D (21) and text-books (16) were searched in order to extract information on drug withdrawals. Monoclonal antibodies and substance combinations were removed from the dataset. Currently, the database comprises two sets of drugs: withdrawn and discontinued. A total of 270 drugs, that were identified to be withdrawn or recalled in at least one country/market due to safety issues are included in the former set while the latter consists of 308 drugs that were suspended or discontinued in at least one market due to unclear reasons. The chemical structures of the withdrawn/discontinued drugs were standardized using the JChem Suite (Instant JChem version 14.10.27.0, ChemAxon (http://www.chemaxon.com)). The standardization steps included aromatization of the structures, addition of explicit hydrogens, removal of salts, and generation of 3D structures. InChIKeys were calculated for the standardized structures and used to join structures from different datasets and to remove duplicates. In addition to InChIKeys, the set was scanned for duplicates using chemical names, canonical smiles and external identifiers.
In many cases, the reason(s) for withdrawal and associated toxicity was directly provided by the source. The reasons were manually extracted for the remaining drugs by performing literature search. Furthermore, the years of first approval, first and last withdrawal, and the year of first reported death for all the withdrawn drugs and most of the discontinued drugs were extracted from the literature. Additionally, the Anatomical Therapeutic Chemical (ATC) codes and external chemical identifiers were collected to link the drugs to the public databases WHO ATC index (http://www.whocc.no/atc_ddd_index/), ChEMBL (22) and PubChem (23), respectively. External identifiers were extracted using the PubChem Identifier Exchange Service (https://pubchem.ncbi.nlm.nih.gov/idexchange/idexchange.cgi) whereas the ATC codes were collected by looking for drug names in the WHO ATC index. For those drugs without an ATC code assigned by the WHO, pseudo-ATC class names were assigned based on their primary indication areas. The acute oral toxicity class was calculated for each drug using the ProTox webserver (24). The toxicity classes (ranging from 1 to 6) are based on the Globally Harmonized System of Classification and Labelling of Chemicals (GHS; https://www.osha.gov/dsg/hazcom/ghs.html) which classifies compounds using their median lethal doses (LD50). Drugs that demonstrated very low structural similarity to the ProTox dataset were assigned to the class 0.
Protein targets
Human protein targets for withdrawn and discontinued drugs were obtained from the Comparative Toxicogenomics Database (CTD) (25) and the ChEMBL database v. 19 (22). The targets from CTD were filtered to obtain only interactions with the interaction types involving activity, binding, transport or metabolic processing. The ChEMBL targets were filtered using the following criteria, adapted from the recommendations on search criteria by Bajorath et al. (26). First, all interactions with an activity comment ‘inactive’, ‘inconclusive’ or ‘not active’ were removed. Second, only interactions with nanomolar (nM) standard units were kept. Third, all interactions with a confidence score below 4 were deleted to remove all non-protein targets. Fourth, only interactions with standard activity relations ‘ = ’, ‘<’, ‘<<’, ‘< =’, ‘ = = ’ and those without a standard activity relation were kept. In the last step, all interactions marked with target types as cell-line and ADMET were omitted to retain only interactions those with protein targets measured in functional or binding assays. As a result, we retained a total of 1.4 million compound-target interactions. Target interactions were assigned to the withdrawn/discontinued drugs by mapping the ChEMBL/CTD compound identifiers which resulted in a total of 20,558 drug-target interactions. These involved 327 drugs and 946 distinct human protein targets. To provide additional information concerning adverse effects, drug-target interactions were classified into therapeutic and potential off-targets. Therapeutic or primary drug targets were identified using mechanism of action information from ChEMBL (22), primary target information from PDB (27), pharmacological action from Drugbank (20) as well as the Therapeutic Target Database TTD (28). Information regarding targets considered as off-targets was gathered from the Novartis Safety Panel list published by Lounkine et al. (29).
Enriched pathways
In order to emphasise the interpretation of drug-target interactions at molecular level, we enriched the biological pathways from ConsensusPathDB (30) using the human protein targets from our database. A total of 149 KEGG pathways were enriched with an enrichment P-value > 0.01 while ensuring that at least two protein targets are involved in each pathway. The 149 enriched pathways comprise different signaling, metabolic and biochemical pathways in addition to the drug-target interaction pathways. Altogether, 703 human protein targets were found to be involved in the enriched pathways.
Genetic variations
Information on genetic variations, or widely known as single nucleotide polymorphisms (SNPs), were extracted from the dbSNP database (31). To extract the SNP information from dbSNP for the human protein targets within our database, the BioMart R package (32) was used. The human genome assembly GRCh38.p3, provided by the Ensembl database (33), was used as a reference genome. SNP information extraction started with a collection of gene symbols or names as defined by the HUGO Gene Nomenclature Committee (HGNC) database (34). The Ensembl-Mart was queried for HGNC symbols and the corresponding Ensembl transcript identifiers were extracted for each gene. The chromosomal position was identified for each transcript and SNP identifiers were used to get additional information including minor allele frequency (MAF) and function predictions from SNP-Mart. This information was mapped to the genes queried for on Ensemble-Mart using the SNP identifiers and transcript identifiers. In order to identify the most important variations, only those SNPs located within the coding region of a protein and marked as missense variants with an MAF value were retained. A total of 889 human protein targets were identified to be associated with 27 790 unique SNP identifiers. In total, 1731 SNPs have a MAF >1%.
Toxicity types
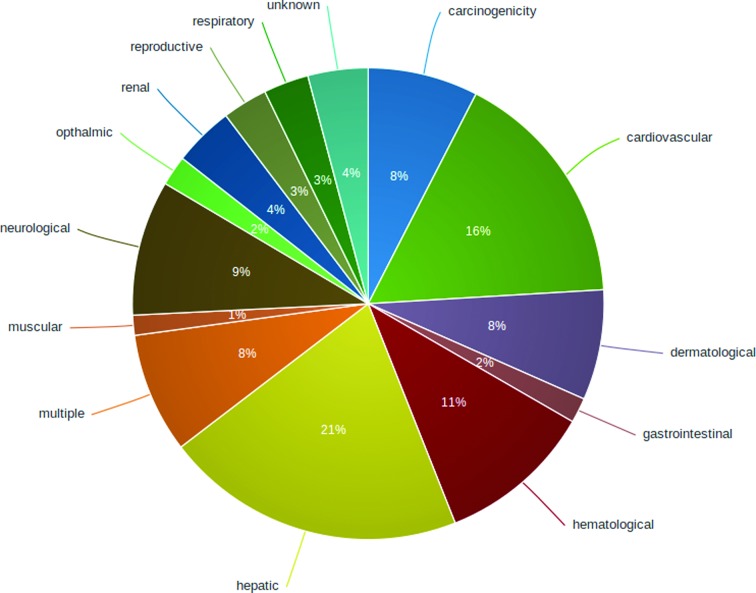
A total of 14 categories of toxicity types were defined based on the adverse effects associated with drug withdrawal. These include the following toxicity types: hepatic, cardiovascular, haematological, dermatological, carcinogenic, neurological, renal, gastrointestinal, ophthalmic, muscular, reproductive and respiratory toxicity as well as the type ‘multiple toxicities’ comprising compounds with observed multiple organ failure as well as ‘unknown toxicity’ where no specific toxic effect could be identified, although a safety issue was associated with the withdrawal. The toxicity types were manually assigned based on the reasons available and also the reasons extracted from the literature. The number of withdrawn/discontinued drugs associated with each toxicity type is summarized in Figure 1 and Supplementary Table S1.
Figure 1.
Overview of toxicity types associated with drug withdrawals.
Server, database and system requirements
WITHDRAWN is based on a relational MySQL database (http://www.mysql.com/). All data is stored on the MySQL database and WITHDRAWN is hosted as a Java web application on a Linux virtual server, accessible at http://cheminfo.charite.de/withdrawn. We strongly recommend using a latest Mozilla Firefox, Google Chrome or Safari browser, with JavaScript options enabled, to access the website.
DATABASE SEARCH OPTIONS
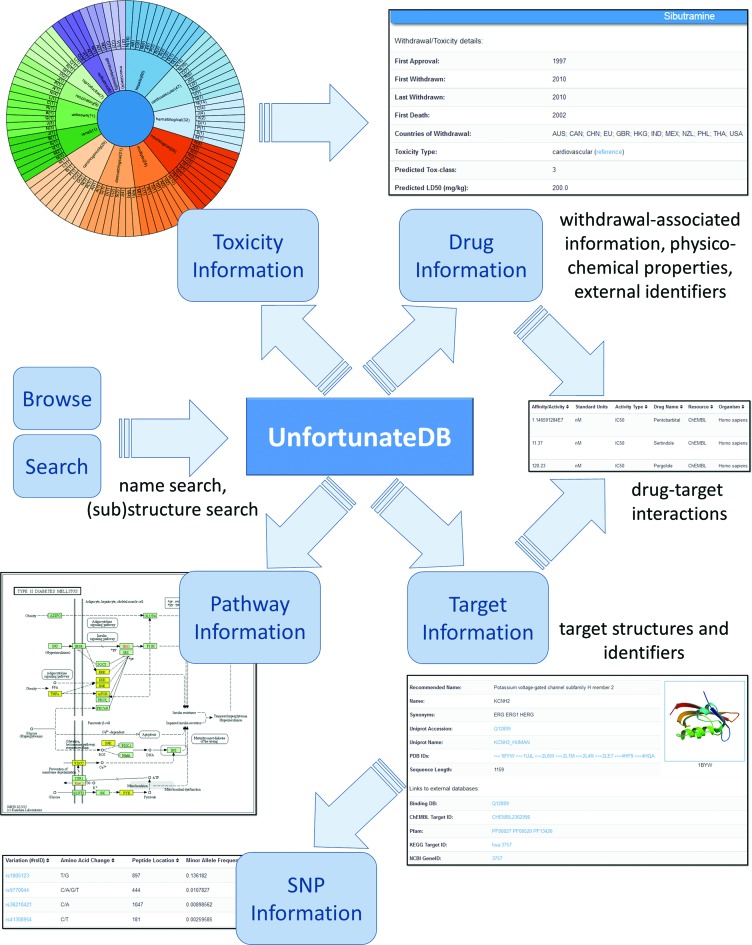
The data presented by WITHDRAWN can be queried via multiple search forms, as summarized in Figure 2. A quick and simple way is to browse through the lists of withdrawn and discontinued drugs. Different search options available on the database include.
Figure 2.
Schematic representation of WITHDRAWN: various search options and different entity types: drugs, targets, pathways, toxicity types and SNPs.
Drug search
Drugs can be searched using multiple options. In case a direct match by name or synonym is not possible, the structure of the queried name is obtained from PubChem and five most similar withdrawn/discontinued drugs will be identified and displayed to the users. When providing a structure input via the molecule sketching tool, the user has the flexibility to search for database compounds at different levels of Tanimoto similarity (fingerprint similarity using MACCS keys) and also to adjust the number of results to be displayed. In addition, a sub-structure search, using Ullmann's algorithm for subgraph isomerism (35), was implemented to provide an option to lookup for withdrawn/discontinued drugs that contain the query structure. Additionally, drugs can be searched using ATC codes. A detailed drug record displays information about drug withdrawal, physicochemical properties and links to external databases. The users can also view the target interactions of the selected drug. Two separate tables for ChEMBL and CTD interactions are displayed. ChEMBL interactions can additionally be filtered using different activity value cutoffs.
Target search
The users can search for protein targets by providing a gene name, UniProt entry number or UniProt entry name (36) as query in the target search form. In addition, it is possible to browse protein targets using their ChEMBL classification. The resulting target record displays various protein identifiers, PDB (http://www.rcsb.org) structures, and links to external target databases. In addition, the interactions of the target with withdrawn/discontinues drugs can be viewed in the same page. The information includes activity types, units and values as well as the organism and information source. Furthermore, the information on biological pathways and SNPs, including amino acid changes, peptide positions, MAFs, PolyPhen scores (37) and links to dbSNP, were added in the detailed record of a target.
Pathway search
To provide clear insights on withdrawn drug-target interaction effects, the pathway maps were extracted from the KEGG database (38,39) for all the enriched biological pathways. In every pathway map, the targets that have an interaction with withdrawn drugs are highlighted. Pathways can be accessed via a selection list. Additionally, the targets highlighted within the map are listed below to provide a link to interacting drugs.
Toxicity type search
Alternatively, the drugs can be browsed by toxicity type. An interactive wheel was designed to visualize different toxicity types using the open source D3 visualization libraries (http://d3js.org/). The users can see number of drugs in each toxicity type as well as the distribution of the drugs into different ATC classes within each toxicity type. Furthermore, the list of drugs classified in each toxicity type can be exclusively viewed by clicking on the toxicity type. Major withdrawal reasons under each toxicity type are summarized in Figure 1 and Supplementary Table S1.
USE CASE
The following use case, represented in Figure 3, illustrates the utility of WITHDRAWN as a knowledge-base to understand the mechanism of adverse drug reactions associated with drug withdrawals:
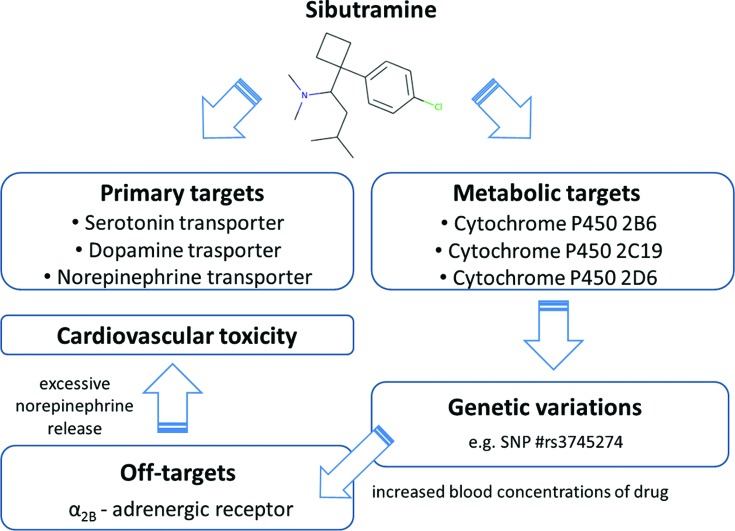
Figure 3.
Case study—use of WITHDRAWN in connecting links between drugs, targets and SNPs in toxicological context.
A search for the drug sibutramine, originally developed by Knoll Pharmaceuticals, as an appetite suppressant for treatment of exogenous obesity reveals that it was recalled in the USA in 2010 due to adverse cardiovascular events including myocardial infarctions and stroke (40). Sibutramine is a non-selective inhibitor that acts by inhibiting the reuptake of the three monoamine neurotransmitters: serotonin, dopamine and norepinephrine. By searching for sibutramine targets in WITHDRAWN, the drug record shows additional drug-target interactions including the cytochromes CYP2B6, CYP2C19 and CYP2D6 as well as the α2B-adrenergic receptor (ADRA2B) where sibutramine exhibits similar activity as at the primary targets. WITHDRAWN shows four genetic variants for CYP2B6 with a MAF above 1% (rs3745274, rs3211371, rs8192709 and rs28399499). Indeed, it has been shown that CYP2B6 variations, particularly rs3745274, may lead to a significant increase in the blood concentration of sibutramine and its active metabolites (41,42). As summarized by Zhang et al. (43), the increased drug concentration could result in an increased off-target activity at ADRA2B which, through an increased norepinephrine release, can lead to increased blood pressure and adverse cardiovascular events. The example emphasizes the importance of considering extensive drug-target and pharmacogenetics studies during drug development.
CONCLUSIONS
WITHDRAWN is a rich resource of withdrawn or discontinued drugs. Due to a relatively small number of drugs withdrawn per year (∼10), we will update the database annually to ensure good coverage and high standard. The database not only contains information related to drug withdrawals and associated adverse drug reactions but also drug-target interactions and genetic variations of the protein targets. The drug-target interaction information is mapped to biological context by enriching the relevant pathways. The illustrated case study proves that, connecting links between drugs, targets and SNPs may explain the underlying mechanisms of toxicity. The knowledge presented in the database can improve the insights of drug-target interactions in toxicological context and provide the rationale for further off-target profiling and enhanced pharmacogenetics studies in different populations.
Acknowledgments
The authors kindly acknowledge Jevgeni Erehman, Mathias Dunkel and Herbert Schulz for their input and support.
Author Contributions: Implementation of website: V.B.S.; curation of drug information: V.B.S., C.O., M.N.D., J.N.; curation of target information: J.N., V.B.S., M.N.D., C.O.; curation of pathway information: V.B.S.; curation of SNP information: B.O.G.; writing of manuscript: mainly V.B.S., input from M.N.D., R.P., J.N., B.O.G.; project coordination: R.P., M.N.D.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Berlin-Brandenburg research platform BB3R, Federal Ministry of Education and Research (BMBF), Germany [031A262C] and the Immunotox project, Federal Ministry of Education and Research (BMBF), Germany [031A268B]. Funding for open access charge: Berlin-Brandenburg research platform BB3R, Federal Ministry of Education and Research (BMBF), Germany [031A262C].
Conflict of interest statement. None declared.
REFERENCES
- 1.DiMasi J.A., Hansen R.W., Grabowski H.G. The price of innovation: new estimates of drug development costs. J. Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 2.Lasser K.E., Allen P.D., Woolhandler S.J., Himmelstein D.U., Wolfe S.M., Bor D.H. Timing of new black box warnings and withdrawals for prescription medications. JAMA. 2002;287:2215–2220. doi: 10.1001/jama.287.17.2215. [DOI] [PubMed] [Google Scholar]
- 3.Podrid P.J. Can antiarrhythmic drugs cause arrhythmia? J. Clin. Pharmacol. 1984;24:313–319. doi: 10.1002/j.1552-4604.1984.tb01840.x. [DOI] [PubMed] [Google Scholar]
- 4.Heijman J., Voigt N., Carlsson L.G., Dobrev D. Cardiac safety assays. Curr. Opin. Pharmacol. 2014;15:16–21. doi: 10.1016/j.coph.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Roy M., Dumaine R., Brown A.M. HERG, a primary human ventricular target of the nonsedating antihistamine terfenadine. Circulation. 1996;94:817–823. doi: 10.1161/01.cir.94.4.817. [DOI] [PubMed] [Google Scholar]
- 6.Giacomini K.M., Krauss R.M., Roden D.M., Eichelbaum M., Hayden M.R., Nakamura Y. When good drugs go bad. Nature. 2007;446:975–977. doi: 10.1038/446975a. [DOI] [PubMed] [Google Scholar]
- 7.Arrowsmith J. Trial watch: phase III and submission failures: 2007–2010. Nat. Rev. Drug Discov. 2011;10:87. doi: 10.1038/nrd3375. [DOI] [PubMed] [Google Scholar]
- 8.Arrowsmith J., Miller P. Trial watch: phase II and phase III attrition rates 2011–2012. Nat. Rev. Drug Discov. 2013;12:569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 9.Thompson R.A., Isin E.M., Li Y., Weaver R., Weidolf L., Wilson I., Claesson A., Page K., Dolgos H., Kenna J.G. Risk assessment and mitigation strategies for reactive metabolites in drug discovery and development. Chem. Biol. Interact. 2011;192:65–71. doi: 10.1016/j.cbi.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Patel H., Bell D., Molokhia M., Srishanmuganathan J., Patel M., Car J., Majeed A. Trends in hospital admissions for adverse drug reactions in England: analysis of national hospital episode statistics 1998–2005. BMC Clin. Pharmacol. 2007;7:9. doi: 10.1186/1472-6904-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards I.R., Aronson J.K. Adverse drug reactions: definitions, diagnosis, and management. Lancet. 2000;356:1255–1259. doi: 10.1016/S0140-6736(00)02799-9. [DOI] [PubMed] [Google Scholar]
- 12.Hussaini S.H., Farrington E.A. Idiosyncratic drug-induced liver injury: an overview. Expert Opin. Drug Saf. 2007;6:673–684. doi: 10.1517/14740338.6.6.673. [DOI] [PubMed] [Google Scholar]
- 13.Ulrich R.G. Idiosyncratic toxicity: a convergence of risk factors. Annu. Rev. Med. 2007;58:17–34. doi: 10.1146/annurev.med.58.072905.160823. [DOI] [PubMed] [Google Scholar]
- 14.Lucena M.I., Andrade R.J., Kaplowitz N., Garcia-Cortes M., Fernandez M.C., Romero-Gomez M., Bruguera M., Hallal H., Robles-Diaz M., Rodriguez-Gonzalez J.F., et al. Phenotypic characterization of idiosyncratic drug-induced liver injury: the influence of age and sex. Hepatology. 2009;49:2001–2009. doi: 10.1002/hep.22895. [DOI] [PubMed] [Google Scholar]
- 15.Greer M.L., Barber J., Eakins J., Kenna J.G. Cell based approaches for evaluation of drug-induced liver injury. Toxicology. 2010;268:125–131. doi: 10.1016/j.tox.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Waller P. Stephens’ Detection of New Adverse Drug Reactions. 5th Edition ed. Chichester, West Sussex, England: John Wiley & Sons, Ltd; 2004. [Google Scholar]
- 17.Onakpoya I.J., Heneghan C.J., Aronson J.K. Delays in the post-marketing withdrawal of drugs to which deaths have been attributed: a systematic investigation and analysis. BMC Med. 2015;13:26. doi: 10.1186/s12916-014-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jefferys D.B., Leakey D., Lewis J.A., Payne S., Rawlins M.D. New active substances authorized in the United Kingdom between 1972 and 1994. Br. J. Clin. Pharmacol. 1998;45:151–156. doi: 10.1046/j.1365-2125.1998.00651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wysowski D.K., Swartz L. Adverse drug event surveillance and drug withdrawals in the United States, 1969–2002: the importance of reporting suspected reactions. Arch. Intern. Med. 2005;165:1363–1369. doi: 10.1001/archinte.165.12.1363. [DOI] [PubMed] [Google Scholar]
- 20.Wishart D.S., Knox C., Guo A.C., Cheng D., Shrivastava S., Tzur D., Gautam B., Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pihan E., Colliandre L., Guichou J.F., Douguet D. e-Drug3D: 3D structure collections dedicated to drug repurposing and fragment-based drug design. Bioinformatics. 2012;28:1540–1541. doi: 10.1093/bioinformatics/bts186. [DOI] [PubMed] [Google Scholar]
- 22.Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M., Kruger F.A., Light Y., Mak L., McGlinchey S., et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Zhou Z., Han L., Karapetyan K., Dracheva S., Shoemaker B.A., et al. PubChem's BioAssay Database. Nucleic Acids Res. 2012;40:D400–412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drwal M.N., Banerjee P., Dunkel M., Wettig M.R., Preissner R. ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014;42:W53–58. doi: 10.1093/nar/gku401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis A.P., Grondin C.J., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B.L., Wiegers T.C., Mattingly C.J. The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y., Bajorath J. Influence of search parameters and criteria on compound selection, promiscuity, and pan assay interference characteristics. J. Chem. Inf. Model. 2014;54:3056–3066. doi: 10.1021/ci5005509. [DOI] [PubMed] [Google Scholar]
- 27.Rose P.W., Prlic A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin C., Zhang C., Zhu F., Xu F., Chen S.Y., Zhang P., Li Y.H., Yang S.Y., Wei Y.Q., Tao L., et al. Therapeutic target database update 2014: a resource for targeted therapeutics. Nucleic Acids Res. 2014;42:D1118–D1123. doi: 10.1093/nar/gkt1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lounkine E., Keiser M.J., Whitebread S., Mikhailov D., Hamon J., Jenkins J.L., Lavan P., Weber E., Doak A.K., Cote S., et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamburov A., Stelzl U., Lehrach H., Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smigielski E.M., Sirotkin K., Ward M., Sherry S.T. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M.H., Baldock R., Barbiera G., et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43:W589–598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullmann J.R. Algorithm for Subgraph Isomorphism. J. ACM. 1976;23:31–42. [Google Scholar]
- 36.Magrane M., Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7, Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James W.P., Caterson I.D., Coutinho W., Finer N., Van Gaal L.F., Maggioni A.P., Torp-Pedersen C., Sharma A.M., Shepherd G.M., Rode R.A., et al. Effect of sibutramine on cardiovascular outcomes in overweight and obese subjects. N. Engl. J. Med. 2010;363:905–917. doi: 10.1056/NEJMoa1003114. [DOI] [PubMed] [Google Scholar]
- 41.Bae S.K., Cao S., Seo K.A., Kim H., Kim M.J., Shon J.H., Liu K.H., Zhou H.H., Shin J.G. Cytochrome P450 2B6 catalyzes the formation of pharmacologically active sibutramine (N-{1-[1-(4-chlorophenyl)cyclobutyl]-3-methylbutyl}-N,N-dimethylamine) metabolites in human liver microsomes. Drug Metab. Dispos. 2008;36:1679–1688. doi: 10.1124/dmd.108.020727. [DOI] [PubMed] [Google Scholar]
- 42.Chung J.Y., Jang S.B., Lee Y.J., Park M.S., Park K. Effect of CYP2B6 genotype on the pharmacokinetics of sibutramine and active metabolites in healthy subjects. J. Clin. Pharmacol. 2011;51:53–59. doi: 10.1177/0091270010362906. [DOI] [PubMed] [Google Scholar]
- 43.Zhang W., Roederer M.W., Chen W.Q., Fan L., Zhou H.H. Pharmacogenetics of drugs withdrawn from the market. Pharmacogenomics. 2012;13:223–231. doi: 10.2217/pgs.11.137. [DOI] [PubMed] [Google Scholar]