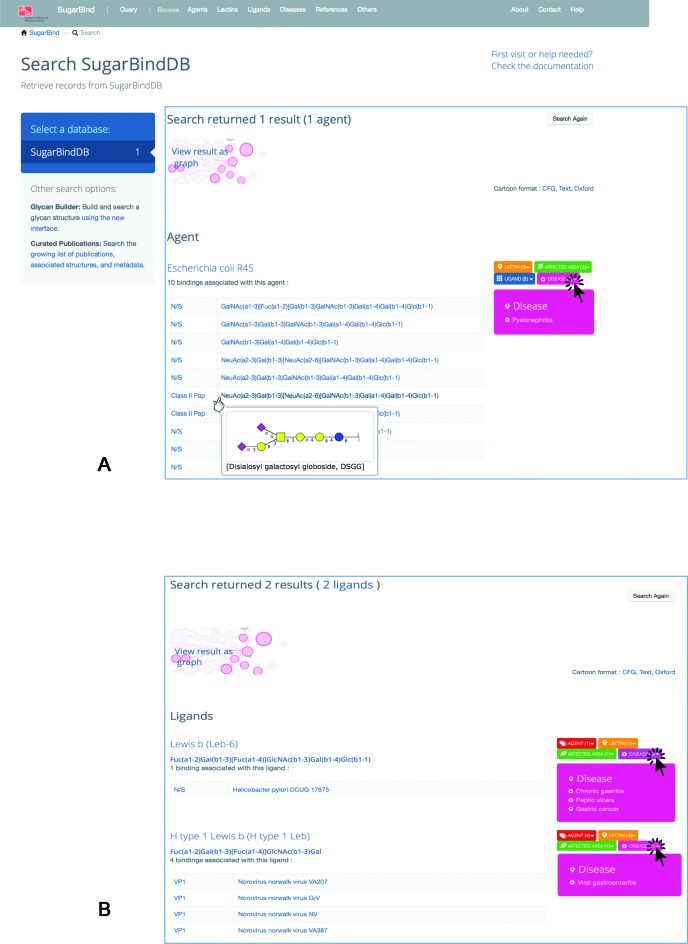
Figure 1.
(A) Result page prompted by querying SugarBindDB with the pathogen/agent name ‘Escherichia coli R45’. The page is organized in blocks as a function of the query. In this case, since the query is an agent, the header is the name of the agent. Then, all bindings reported in the database for this agent are shown. The glycan molecule is presented linearly for the sake of simplicity but mousing over the formula prompts a two-dimensional cartoon presenting the Essentials of Glycobiology 2nd Edition/CFG nomenclature symbols. The association boxes are displayed on the right side with a colour code corresponding to the entity type, orange for lectins, red for agents, green for affected area/tissue, pink for disease. Clicking on any of these boxes prompts the list of entities sharing the same binding. The screen capture shows the effect of clicking on disease. (B) Result page prompted by querying SugarBindDB with the ligand sequence: Fuc(a1–2)Gal(b1–3)[Fuc(a1–4)]GlcNAc(b1–3)Gal. In this case, the header is the name of the ligand and two answers are returned. It is shown here that two unrelated pathogens a bacterium (H. pylori) and a virus (Norovirus Norwalk) recognize a similar glycan ligand.