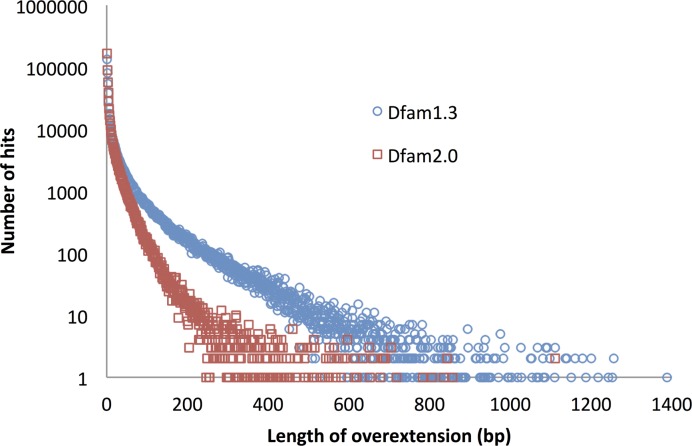
Figure 3.
Distribution of overextension lengths. Profile HMMs for human Dfam families were searched against an overextension benchmark trained on human sequence data, built using GARLIC. For each hit above GA threshold, overextension was calculated. The plot shows, for each overextension length, the number of hits with that length. Application of our two changes (increased average relative entropy and exponential entropy weighting) clearly reduced the frequency of very long overextensions.